Method for increasing amount of metabolites in fermentation cells and preparing IDMS standard
A standard product and metabolite technology, applied in the field of microbial fermentation, can solve the problems of hindering the metabolic engineering research process of metabolites, low intracellular metabolite concentration, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Embodiment 1, fermentation and substrate add
[0078] The fermenter uses a 1L tank produced by Shanghai Guoqiang Biochemical Engineering Equipment Co., Ltd. The volume of the fermentation broth is 0.6L, and the inoculum volume is 1%. The seeds are pre-cultivated at 220rpm and 30°C for 24h, and then 6mL is centrifuged to remove Add 6mL normal saline to redissolve the supernatant, put it into the fermenter, and use OD 600 Test bacteria concentration. The whole process uses NaOH solution to remove CO from the aeration 2, the ventilation rate is 0.6L / min (1vvm), the stirring speed: the initial fermentation group of the substrate supplementation experiment is about 300rpm, and the subsequent 400rpm; the fermentation group without the substrate supplementation experiment is 400rpm, and the fermentation temperature is controlled at 30°C , use ammonia water to control the pH at 5.0, and maintain the tank pressure at 0.05MPa. For the whole fermentation process, the Biostar so...
Embodiment 2
[0082] Embodiment 2, macro data analysis of fermentation process
[0083] Fermentation was carried out by the method of Example 1, and data comparison analysis was performed on the groups without substrate supplementation experiment and the substrate supplementation experiment group. The results are shown in Table 4.
[0084] Table 4
[0085]
[0086]
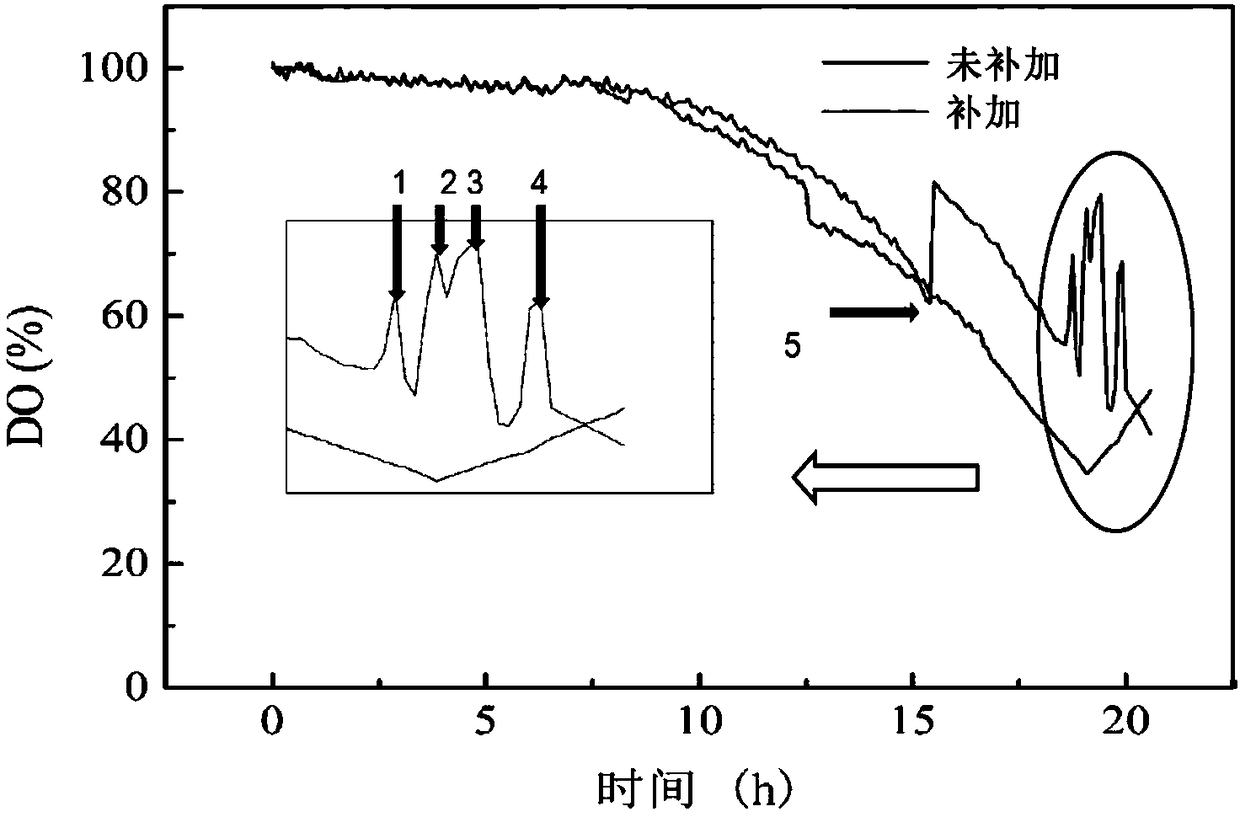
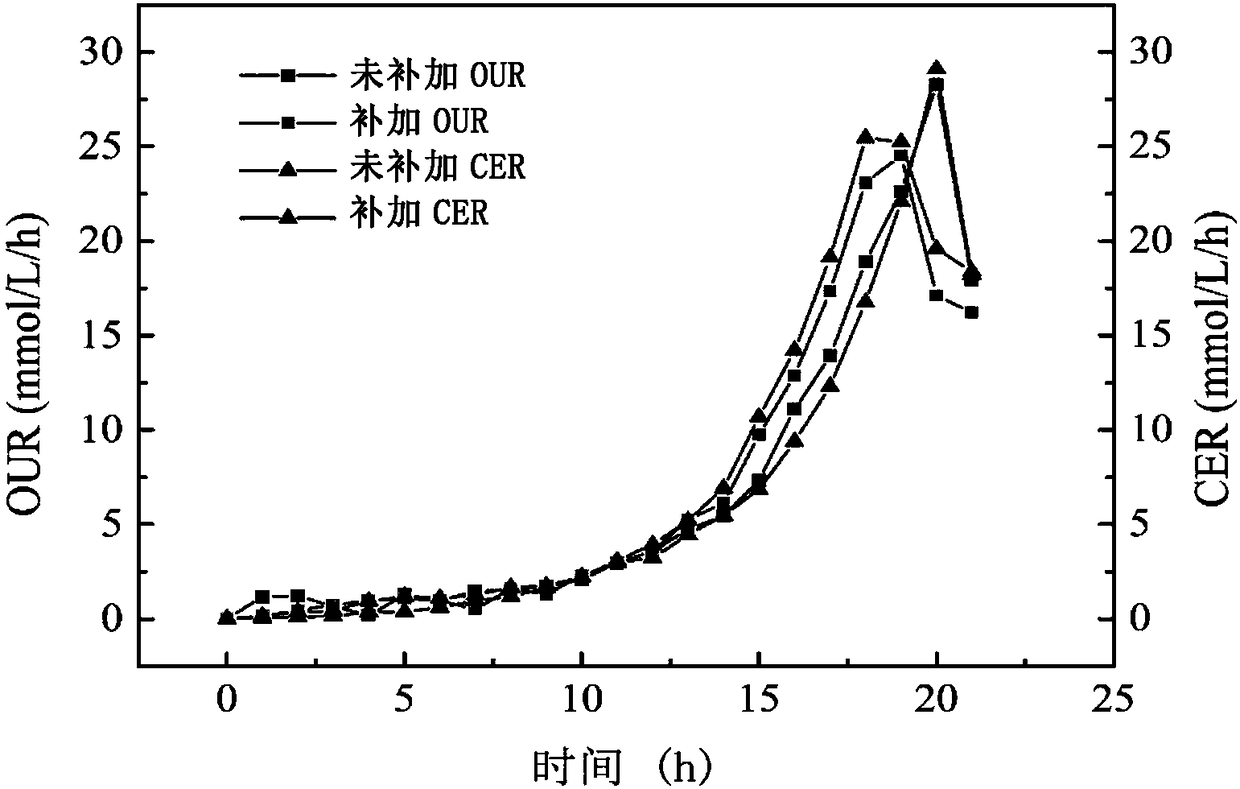
[0087] The results show that due to the large culture volume, the post-processing time of a single sampling sample is long. In order to minimize the contact time between the sample and cold methanol and prevent a large amount of intracellular metabolites from leaking, add it in 4 times and add it in a single time. 13 The way of C full-standard glucose concentration is 1.5g / L is relatively most suitable. When the dissolved oxygen DO starts to rise and the OUR starts to drop 13 C Add fully labeled glucose, start rapid sampling operation after 5 minutes, each sampling 0.15L. When the oxygen uptake rate (OUR) and carbon d...
Embodiment 3
[0090] Embodiment 3, the acquisition of IDMS standard product
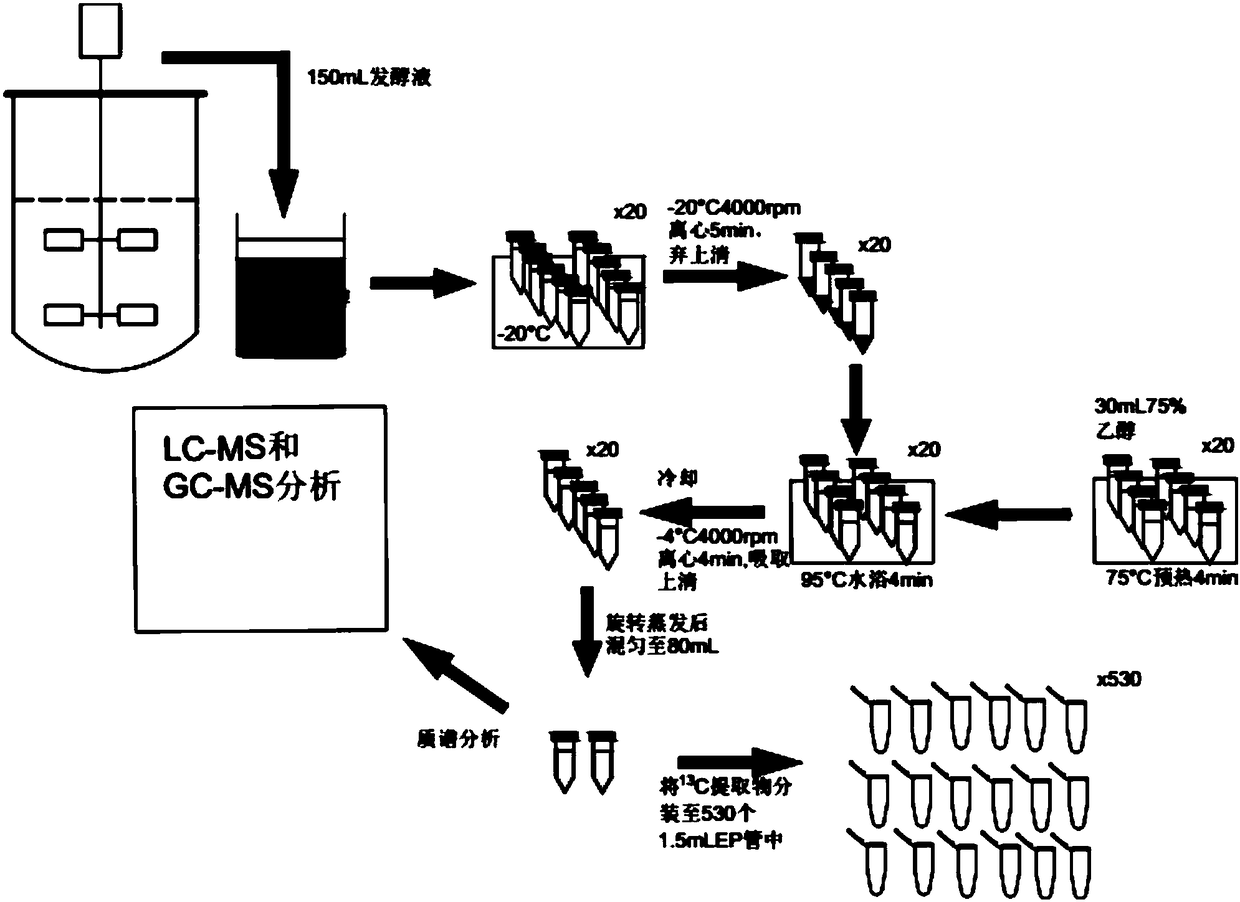
[0091] The method of Example 1 was used to carry out fermentation and substrate supplementation stimulation treatment (supplementation mode 3), and the fermentation broth was extracted to prepare IDMS standard substance. The specific method for preparing IDMS standard items is as follows: figure 1 shown.
[0092] Standards are quantified by known concentration 12 C standard quantitation unknown 13 C metabolite concentration. separate a certain amount of 12 C Phosphate Sugar Standards, 12 C standard organic acids, 12 C nucleotide standards and 12 C Amino Acid Standards Made 4 Kinds of Mixed Standards, 12 The concentration range of standard C is 50-0.05 μmol / L except for amino acids, and the concentration range of amino acids is 200-0.1 μmol / L, and a series of concentration gradients are taken. Finally, the different concentration gradients 12 C mixed standard with 13 C Intracellular metabolites are mixed...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com