DNA polymerase, nucleic acid test method and nucleic acid test kit
A polymerase and kit technology, applied in the field of nucleic acid detection, can solve the problems of high RNA template ratio requirements, unsatisfactory reverse transcription amplification efficiency, etc., and achieve the effects of short detection time, low cost and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
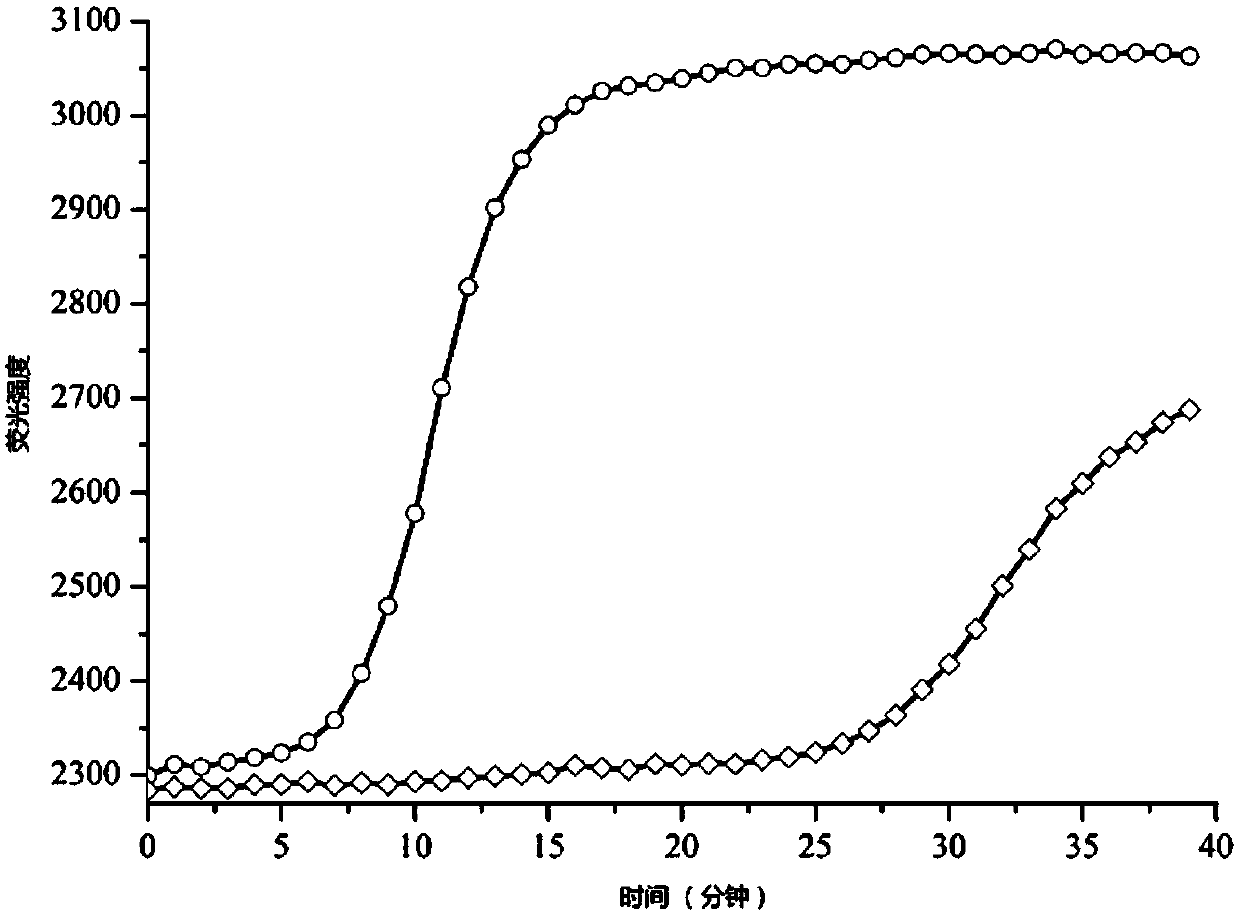
[0091] Embodiment 1 DNA polymerase of the present invention and the activity of former Klenow large fragment
[0092] In this example, the activities of the modified Klenow large fragment (MT-Klenow) and the wild-type Klenow large fragment (WT-Klenow) were compared (that is, the amino acid substitutions described herein were introduced). Using the same RNA template and primers, MT-Klenow and WT-Klenow were used to perform isothermal nucleic acid amplification under the same conditions. The RNA template sequence used is:
[0093] CAGGGAGGUGCCUUGAUGACAUAGAAGAAGAACCAGAUGAUGUUGAUGGCCCAACUGAAAUAGUAUUAAGGGACAUGAACAACAAAGAUGCAAGGCAAAAGAUAAAGGAGGAAGUAAACACUCAGAAAGAAGGGAAGUUCCGUUUGACAAUAAAAAGGGAUAUGCGUAAUGUAUUGUCCCUGAGAGUGUUAGUAAACGGAACAUUCCUCAAACACCCCAAUGGAUACAAGUCCUUAUCAACUCUGCAUAGAUUGAAUGCAUAUGACCAGAGUGGAAGGCUUGUUGCUAAACUUGUUGCUACUGAUGAGCUUACAGUGGAGGAUGAAGAAGAUGGCCAUCGGAUCCUCAAUUCACUCUUCGAGCGUCUUA(SEQ ID NO:19);
[0094] The forward primer sequence is CAGGGAGGTGCCTTGATGACATAGAAGAAG...
Embodiment 2
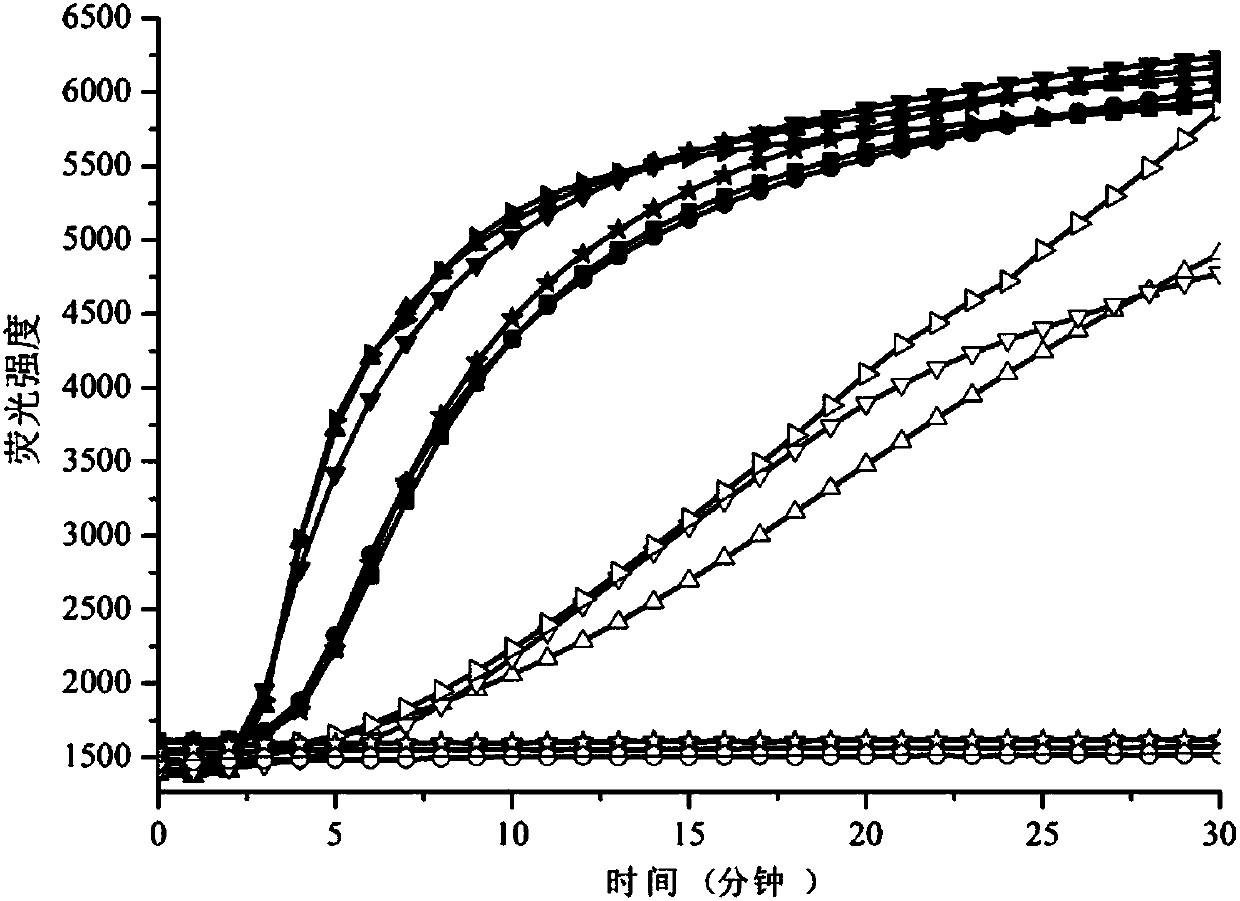
[0100] Embodiment 2: comparative experiment of stem-loop structure primer and linear primer
[0101] The 3' end of the stem-loop structure primer of the present invention is completely identical or completely complementary to the nucleic acid target sequence; an additional 2-15 nt base is added to the 5' end of the primer to form a stem-loop structure complementary to the 3' end of the primer; or 5' The base of 2-15nt is added to the end, and it is complementary to the base of 1-10nt from the 3' end to form a stem-loop structure with a foothold.
[0102] According to the above design principles of stem-loop primers, the present invention compared the isothermal amplification reactions of a pair of common linear primers and their stem-loop primers experimentally. 其中DNA模板序列为:5'-GACAGACTGCACAGGGCATGGATTACTTACACGCCAAGTCAATCATCCACAGAGACCTCAAGAGTAATAATATATTTCTTCATGAAGACCTCACAGTAAAAATAGGTGATTTTGGTCTAGCTACAGAGAAATCTCGATGGAGTGGGTCCCATCAGTTTGAACAGTTGTCTGGATCCATTTTGTGGATGGCACCAGAAGTCATCAG...
Embodiment 3
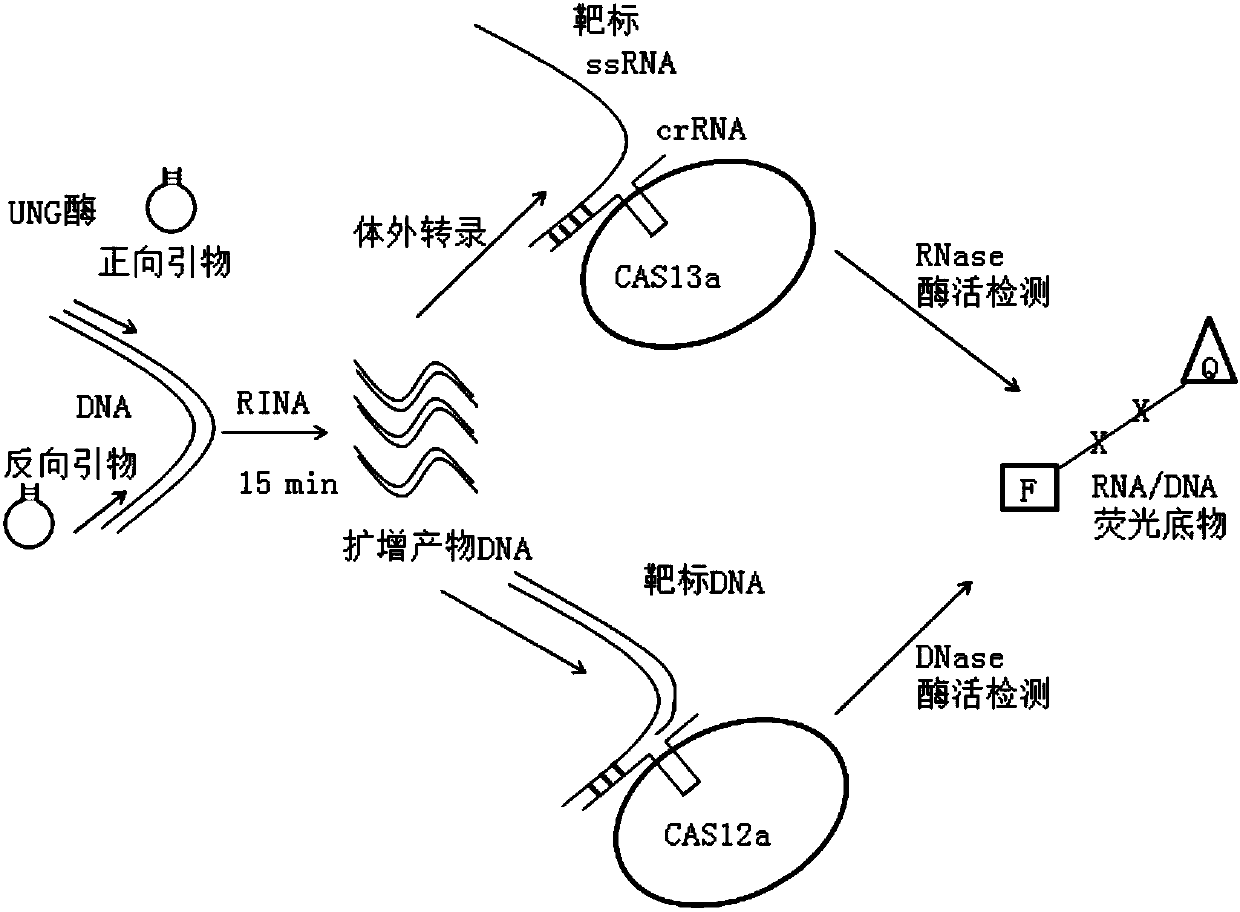
[0106] Example 3 utilizes RINA-CAS (using LwCas13a) technology to detect samples containing influenza B virus (Influenza B)
[0107] First, nucleic acid was extracted from the Influenza B (subtype B influenza virus yamagata) sample, and the nucleic acid extraction kit was Qiagen's viral RNA extraction kit (QIAamp Viral RNA Mini Kit). A sample without Influenza B was used as a negative control.
[0108] Take 1 μL of the extracted RNA and add it to the nucleic acid amplification reaction system at a concentration of 10 -5 Two amplification primers of M Influenza Primer F (sequence is: 5'- ACCAACTTAATACGACTCACTATAGGTGAAACTGCGGTGGGAGTCTTATCCCAAGTTGGT-3' (SEQ ID NO: 6)), Influenza Primer R (sequence: 5'- TGGTTG TCACAAGCACTGCCTGCTGTACACTTCAACCA-3' (SEQ ID NO: 7)) each 2.4 μL, incubated at 37°C for 15 minutes. The designed primers were directed against the conserved gene InfluenzaB NS1 of the yamagata subtype of influenza B virus. The volume of the amplification reaction system...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com