Method for screening miR-181b target gene
A mir-181b, target gene technology, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
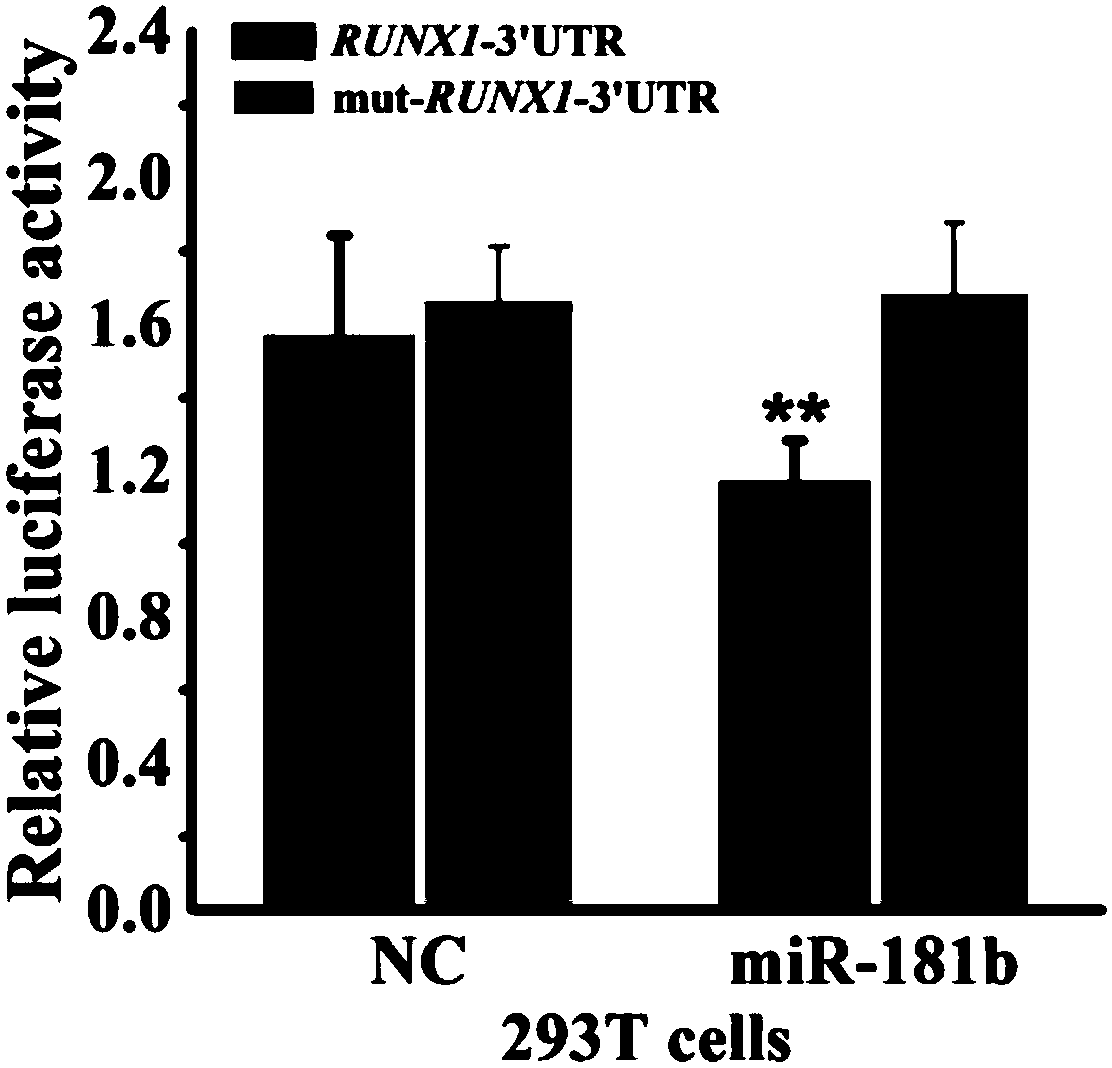
[0065] This embodiment provides a method for screening chi-miR-181b target gene, comprising the following steps:
[0066] 1) Using goat ovary tissue cDNA as template, in Taq DNA polymerase, buffer environment, Mg 2+ 1. In the presence of dNTPs, use the primer RUNX1 to amplify under PCR conditions, and determine that the obtained PCR product is the sequence of the 3'UTR region of the RUNX1 gene;
[0067] The primer RUNX1 is as follows (the underline is the restriction site of XhoI and NotI endonuclease):
[0068] Upstream primer F: 5'-CG CTCGAG AGATCAGTATTGACGCTGATCA-3';
[0069] Downstream primer R: 5'-AT GCGGCCGC CATCCAGAATATCACAAATAACC-3'.
[0070] The condition of described PCR amplification is:
[0071] 15 μL reaction system, including 0.5 μL DNA template, 7.5 μL MasterMix, 0.5 μL upstream and downstream primers, add sterilized water to 15 μL;
[0072] Described PCR amplification reaction procedure is as follows:
[0073] The PCR reaction program using primer RUNX1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com