Long-chain non-coding RNA for early diagnosis of human prostate cancer and its preparation and use
A long-chain non-coding, prostate cancer technology, applied in the direction of DNA preparation, DNA / RNA fragments, recombinant DNA technology, etc., can solve the problems of low correlation and low diagnostic accuracy of prostate cancer, and achieve high correlation and step-by-step Simple, Accuracy-Enhancing Effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
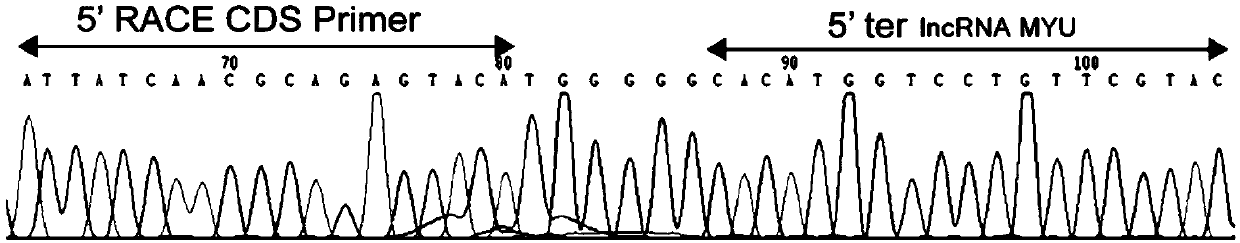
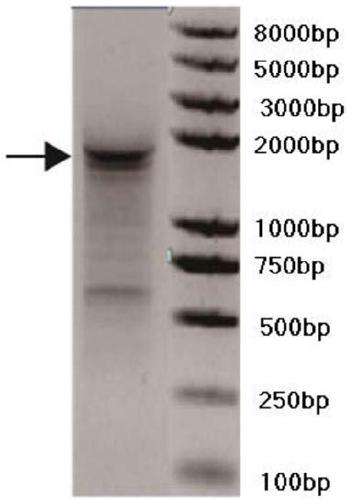
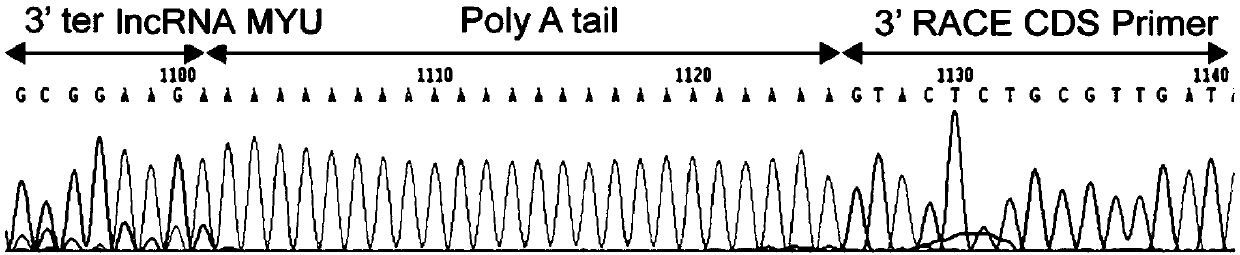
[0082] Example 1 RACE method to obtain precise full-length cDNA sequence of lncRNA MYU
[0083] This embodiment provides a method for obtaining the precise full-length cDNA sequence of lncRNA MYU, which is the long-chain non-coding RNA of the present invention, which specifically includes the following steps:
[0084] 1. Use the extraction reagent RNAiso Plus reagent (Takara) to extract the total RNA of the prostate cancer cell line DU145 by the Trizol extraction method, and use Thermo Fisher's PrimerScript RT-PCR kit (Takara) reverse transcription kit to use oligo dT as reverse transcription Primer, reverse transcription of total RNA into cDNA.
[0085] 2. Using the cDNA in step 1 as a template, design primers 5'-F and 3'-R according to the predicted sequence (the predicted sequence is the known sequence Assembly Gene ID in the Mitranscriptome database: G032453), and the primer 5' The nucleotide sequence of -F is shown in SEQ ID NO: 10, and the nucleotide sequence of the primer 3'-...
Embodiment 2
[0093] Analysis of the conservation, transcription activity and coding ability of the long non-coding RNA described in Example 2
[0094] In this embodiment, bioinformatics algorithms are used to analyze the conservation, transcription activity, and coding ability of the long non-coding RNA (precise full-length cDNA sequence of lncRNAMYU) obtained in Example 1. The specific analysis method is as follows:
[0095] 1. Use Roadmap to obtain the data of H3K4me3, H3k36me3 and H3K27ac in the prostate cancer cell line PC3, and use the ENCODE database (https: / / www.encodeproject.org / ) to obtain the data of H3K4me3, H3k36me3 and H3K27ac in normal tissues, GEO accession number They are GSE96019 (H3K4me3, PC3), GSE96418 (H3k36me3, PC3) and GSE96399 (H3K27ac, PC3), ENCODE Experiment IDs are ENCSR748RBT (H3K4me3, Normal prostate), ENCSR499FXI (H3k36me3, Normal prostate ID) and ENCSR 76327 (Normal prostateID) , And display the obtained data on the UCSC website (http: / / genome.ucsc.edu / ) to obtain ...
Embodiment 3
[0098] Example 3 Detection of the long-chain non-coding RNA expression in different prostate cancer cell lines
[0099] This embodiment provides a specific primer for detecting the expression level of the long non-coding RNA (lncRNA MYU) in cells and tissues; the specific primer includes the upstream primer MYU- F and the downstream primer MYU-R, the nucleotide sequence of the upstream primer MYU-F is shown in SEQ ID NO. 4, and the nucleotide sequence of the downstream primer MYU-R is shown in SEQ ID NO. 5.
[0100] This embodiment provides a kit for early diagnosis of human prostate cancer, which includes the above-mentioned specific primers.
[0101] The kit for early diagnosis of human prostate cancer includes: Premix Ex Taq (TliNaseH Plus) (2×), 10 μL; the upstream primer MYU-F, 10 μM, 0.4 μL; the downstream primer MYU-R, 10 μM, 0.4 μL; ROX Reference Dye II, 0.4 μL; dH 2 O, 6.8 μL.
[0102] This embodiment provides a qRT-PCR reaction system for early diagnosis of human prostate ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com