Binding solution for purification of nucleic acid and application of binding solution
A nucleic acid and buffer technology, applied in the field of purifying nucleic acid binding solution, can solve problems to be improved, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
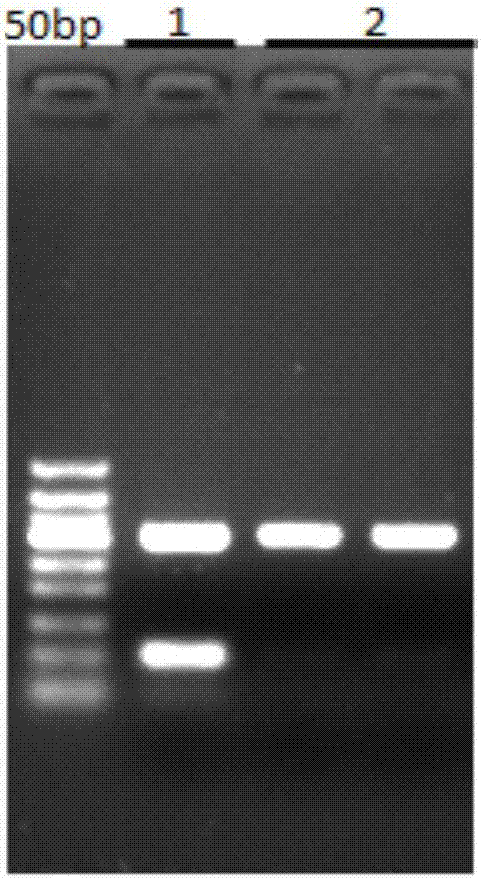
[0039] A 200bp non-specifically amplified fragment was removed from a 2kb polymerase chain reaction (PCR) product. The specific operation steps are as follows:
[0040] (1) Add 0.6 times the volume of binding solution (the binding solution contains 2.5mol / L potassium chloride, 20V% tetraethylene glycol and 100mMTRis, and the pH value is 7.5) and 5 μl of 1 μm average particle size to 40 μl of the sample to be recovered and Surface-modified magnetic beads with carboxyl functional groups, mixed thoroughly, and oscillated for 5 minutes to facilitate the binding of the magnetic beads to the target fragment;
[0041] (2) Place the centrifuge tube in step (1) on the magnetic stand for 2 minutes, and carefully remove the liquid when the magnetic beads are completely absorbed;
[0042] (3) Place the centrifuge tube in step (2) on the magnetic stand, add 300 μl of rinsing solution (the rinsing solution contains 75% ethanol), and do not disturb the magnetic beads;
[0043] (4) Place th...
Embodiment 2
[0050] The 100 bp non-specific amplification band was removed from the 300 bp polymerase chain reaction (PCR) product. The specific operation steps are as follows:
[0051] (1) Add 1 volume of binding solution (the binding solution contains 3.5mol / L sodium chloride, 30V% polyethylene glycol 4000 and 100mM3-MOPS, pH value is 7.0) and 5μl average particle size to 50μl of the sample to be recovered. Magnetic beads with a diameter of 1 μm and surface-modified carboxyl functional groups were mixed thoroughly and shaken for 5 minutes to facilitate the binding of the magnetic beads to the target fragment;
[0052] (2) Place the centrifuge tube in step (1) on the magnetic stand for 2 minutes, and carefully remove the liquid when the magnetic beads are completely absorbed;
[0053] (3) Place the centrifuge tube in step (2) on a magnetic stand, add 300 μl of rinsing solution (the rinsing solution contains 70% ethanol), and do not disturb the magnetic beads;
[0054] (4) Place the cent...
Embodiment 3
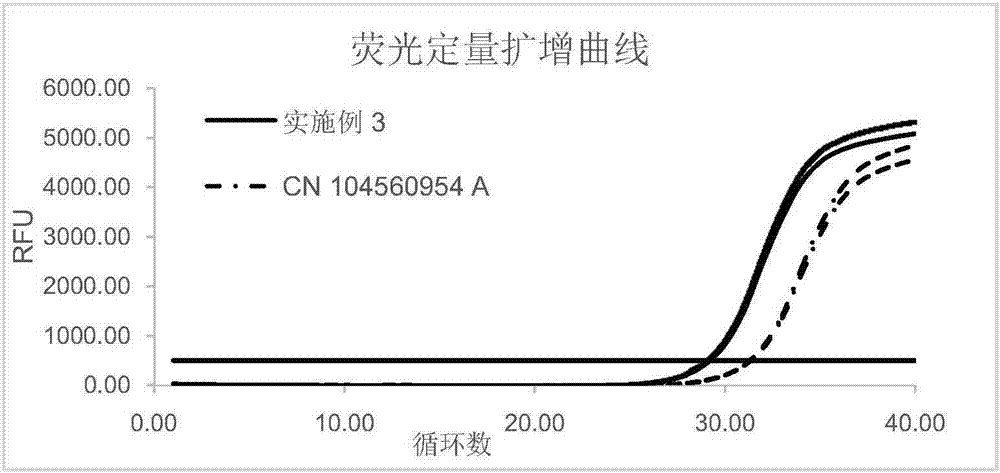
[0061] Purify and recover the small amount of circulating nucleic acid after the linker is added during the construction of the next-generation sequencing library. The specific operation steps are as follows:
[0062] (1) Add 50 μl of binding solution (the binding solution contains 3.5M potassium chloride, 25 mass % PEG6000 and 100 mMT Ris, pH value is 9) and 5 μl of the product with an average particle size of 1 μm and surface modification For magnetic beads with carboxyl functional groups, shake for 5 minutes to facilitate the full binding of the magnetic beads to the target fragment;
[0063] (2) Place the centrifuge tube in step (1) on the magnetic stand for 2 minutes, and carefully remove the liquid when the magnetic beads are completely absorbed;
[0064] (3) Place the centrifuge tube in step (2) on a magnetic stand, add 300 μl of rinsing solution (the rinsing solution contains 80% ethanol), and do not disturb the magnetic beads;
[0065] (4) Place the centrifuge tube ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| The average particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com