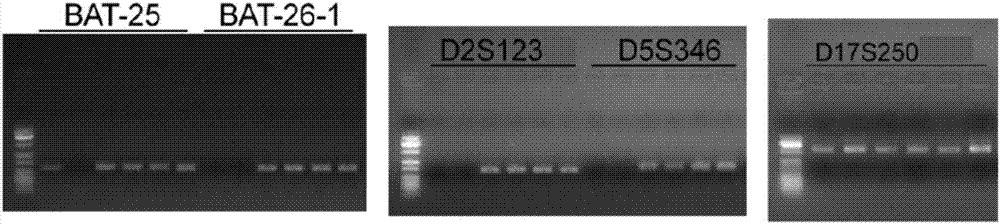
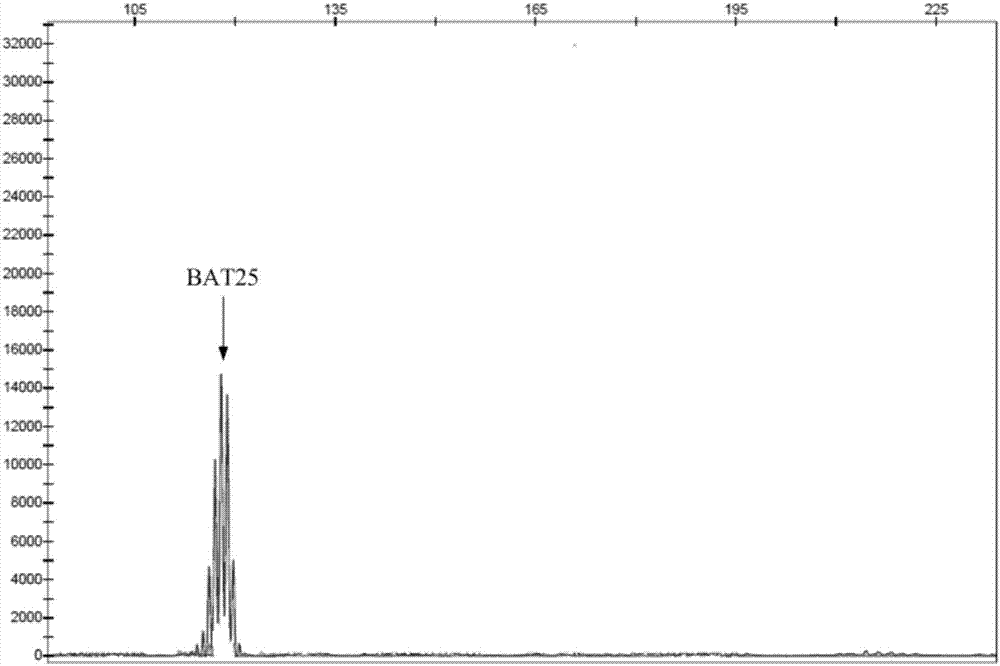
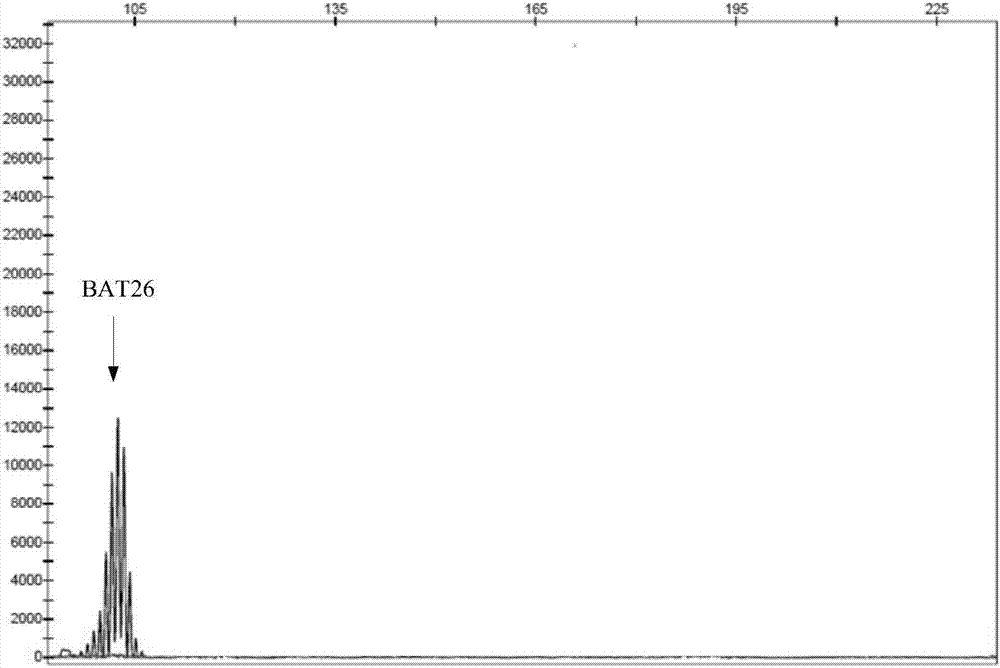
Multiple-fluorescent PCR (polymerase chain reaction) amplification reagent and kit for detecting instability of microsatellite
An unstable, multiple fluorescence technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of easy cross-overlapping of fluorescence peaks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Check the gene number of the microsatellite site to be tested in the Gene Bank, and determine the main repeat sequence of each microsatellite site and the base sequence of about 1000 bp upstream and downstream of the main repeat sequence. The relevant information of the microsatellite site is shown in Table 1 shown.
[0078] Table 1: Basic information of microsatellite loci
[0079] Microsatellite loci
gene number
major repeat sequence
BAT25
L04143
(A) 25
BAT26
U41210
(A) 26
D2S123
Z16551.1
(CA) 15
D5S346
NM_005669
(CA) 20
D17S250
NR_033753.2
(CA) 24
[0080] Targeted design of upstream primers and downstream primers for each microsatellite locus, and correspondingly labeled fluorescent groups, as shown in Table 2.
[0081] Table 2: Upstream and downstream primers for microsatellite loci
[0082]
[0083]
[0084] The designed primers were biosynthesized by Sangon,...
Embodiment 2
[0086]The upstream primer BAT25-F1, downstream primer BAT25-R1, upstream primer BAT26-F1, downstream primer BAT26-R1, upstream primer D2S123-F1, downstream Primer D2S123-R1, upstream primer D5S346-F1, downstream primer D5S346-R1, upstream primer D17S250-F1 and downstream primer D17S250-R1 are the same as in Example 1, the difference is that microsatellites for the control are also added in this example The upstream primer Penta C-F1 and the downstream primer Penta C-R1 designed at site Penta C. The relevant information of the microsatellite locus Penta C is shown in Table 3.
[0087] Table 3: Basic information of microsatellite locus Penta C
[0088] Microsatellite loci
gene number
major repeat sequence
Penta C
AL138752
(AAAAG) 3
[0089] The upstream primer Penta C-F1 and the downstream primer Penta C-R1 designed by Penta C were specifically designed for the microsatellite site Penta C, and correspondingly labeled fluorophores, as shown...
Embodiment 3
[0095] The upstream primer BAT25-F1, downstream primer BAT25-R1, upstream primer BAT26-F1, downstream primer BAT26-R1, upstream primer D2S123-F1, downstream Primer D2S123-R1, upstream primer D5S346-F1, downstream primer D5S346-R1, upstream primer D17S250-F1 and downstream primer D17S250-R1 are the same as in Example 1, the difference is that microsatellites for the control are also added in this example The upstream primer Penta D-F1 and the downstream primer Penta D-R1 designed at site Penta D. The relevant information of the microsatellite locus Penta C is shown in Table 5.
[0096] Table 5: Basic information of microsatellite loci Penta D
[0097] Microsatellite loci
gene number
major repeat sequence
Penta D
Ac000014
(AAAAG) 8
[0098] The upstream primer Penta D-F1 and the downstream primer Penta D-R1 designed by Penta D were specifically designed for the microsatellite site Penta D, and correspondingly labeled fluorescent groups, as...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com