Detection chip for tumor driver gene and its application
A technology for detecting chips and driver genes, applied in the biological field, can solve the problems of difficult detection and low sensitivity of tumor driver genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
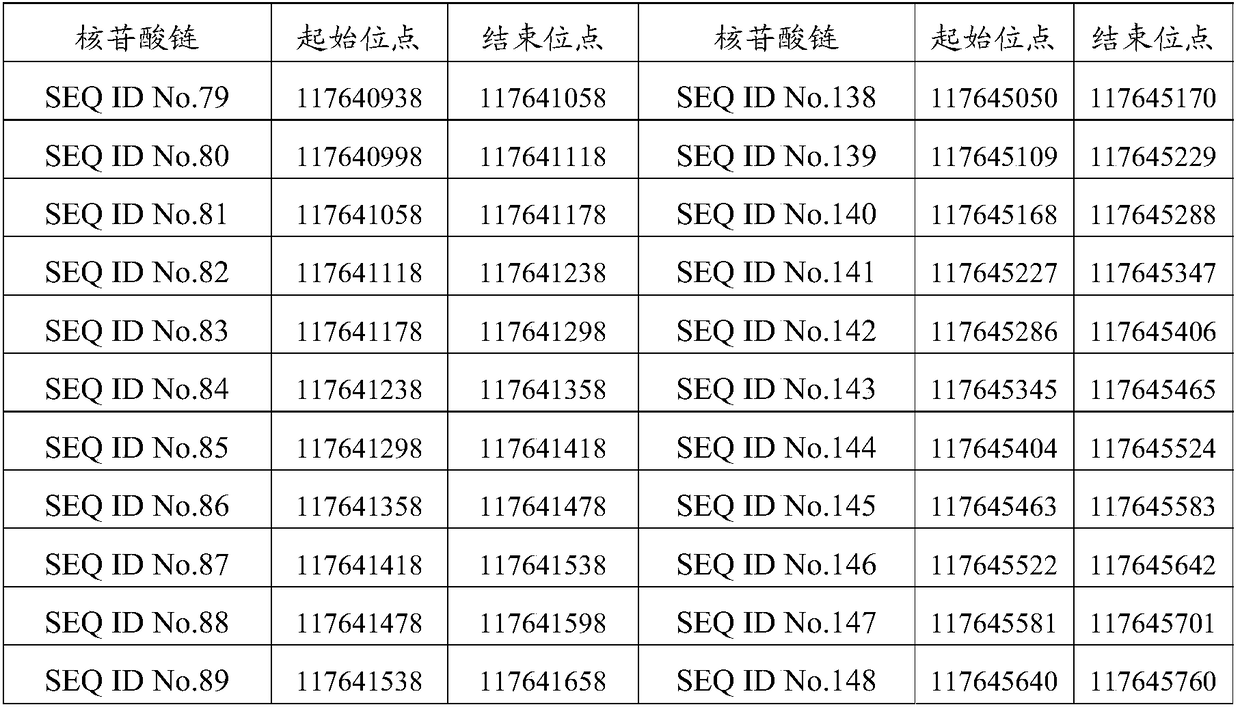
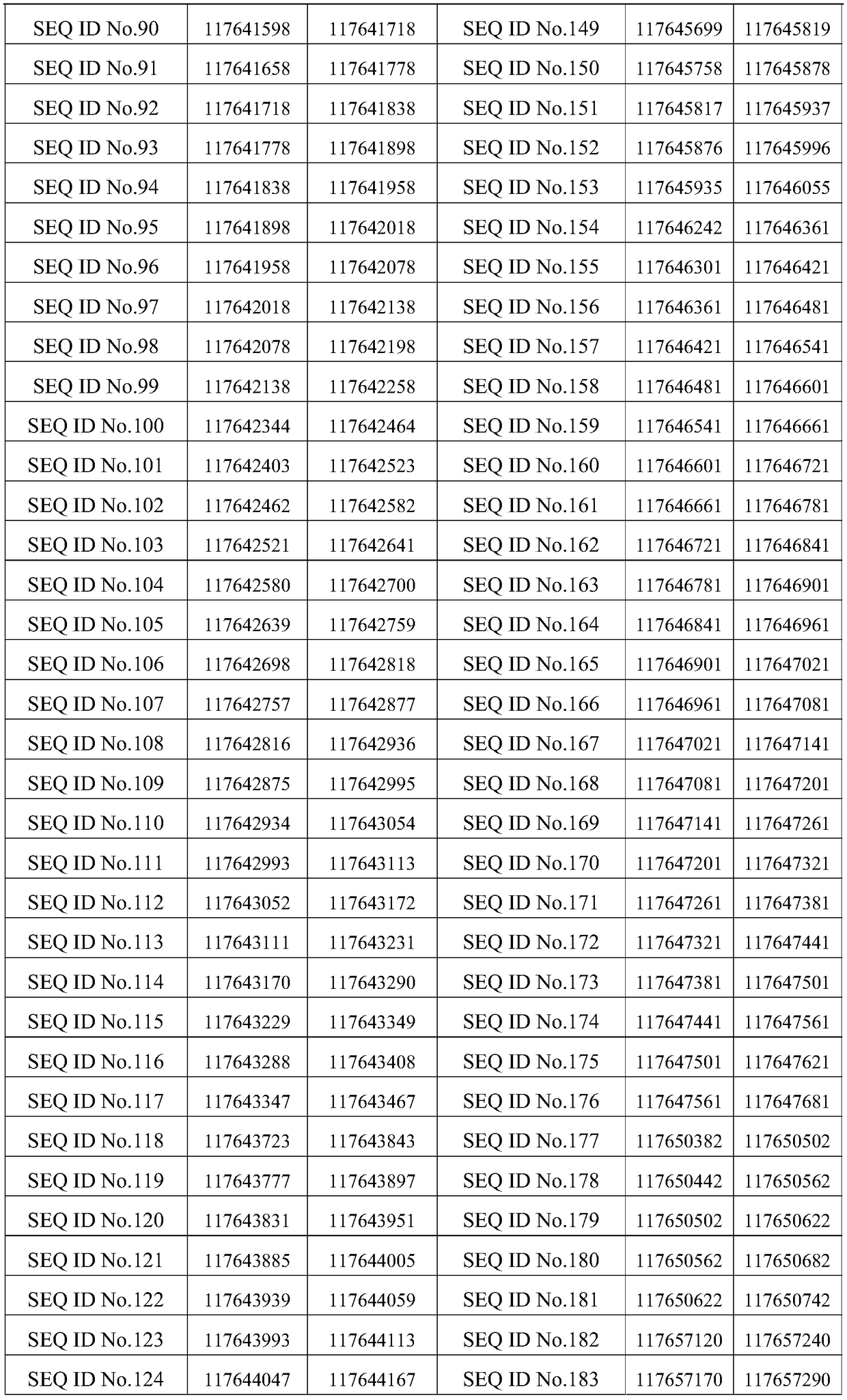
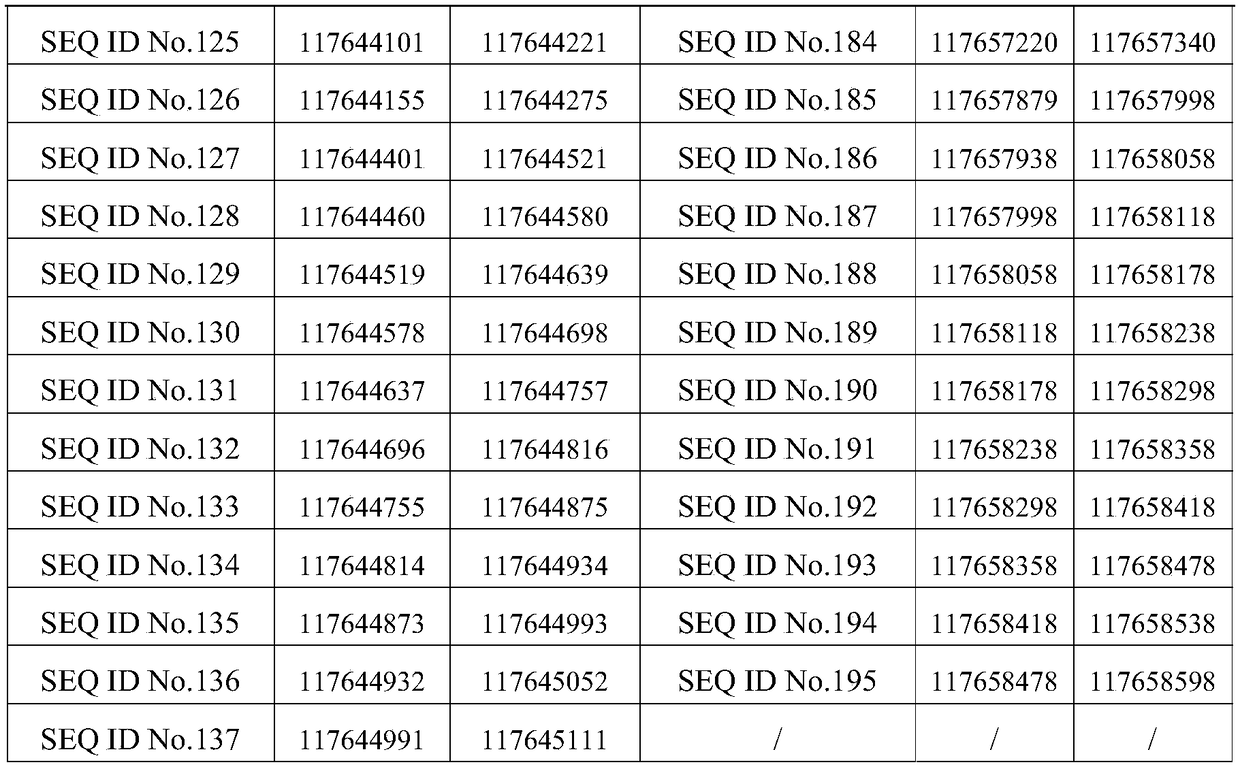
[0150] 236 probes of sequences shown in SEQ ID No.1 to SEQ ID No.236 are arranged on the detection chip, and the venous blood of patients with non-small cell lung cancer is detected as 12 samples (provided by Jinhua Hospital of Traditional Chinese Medicine) with the detection chip. The 12 tumor driver genes are ALK, BRAF, DDR2, EGFR, ERBB2, FGFR1, KRAS, MET, NRAS, PIK3CA, RET and ROS1. The sequencing platform is Illumina, the reference genome is GRCh37 / hg19, and the reference transcriptome is Ensembl75. The reference databases are COSMIC (version v74), dbSNP (version v142), iCMDB (version 4.0.0). The filter conditions are as follows:
[0151] 1. Keep the variants that meet the following filter conditions: a. Base quality > 20, b. Sequence alignment quality > 50, c. Strand preference = 30, f. The number of sequencing short fragments supporting SNP variation in the sample >= 3, g. The number of sequencing short fragments supporting Indel variation in the sample >= 5.
[0152] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com