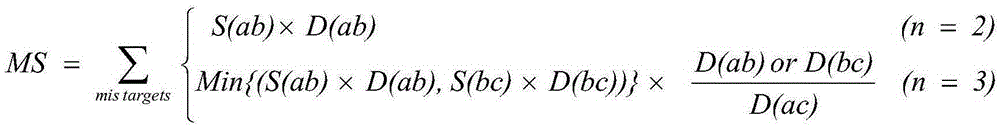CRISPR-Cas9 system sgRNA action target screening method and apparatus
A target and function technology, applied in the field of screening of sgRNA targets in the CRISPR-Cas9 system, can solve the problems of lack of mature methods for genome-wide targets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 Screening method for the sgRNA target of the CRISPR-Cas9 system designed for chicken
[0060] In this example, chicken, a representative animal of poultry, is taken as an example to design a genome-wide Cas9 target library.
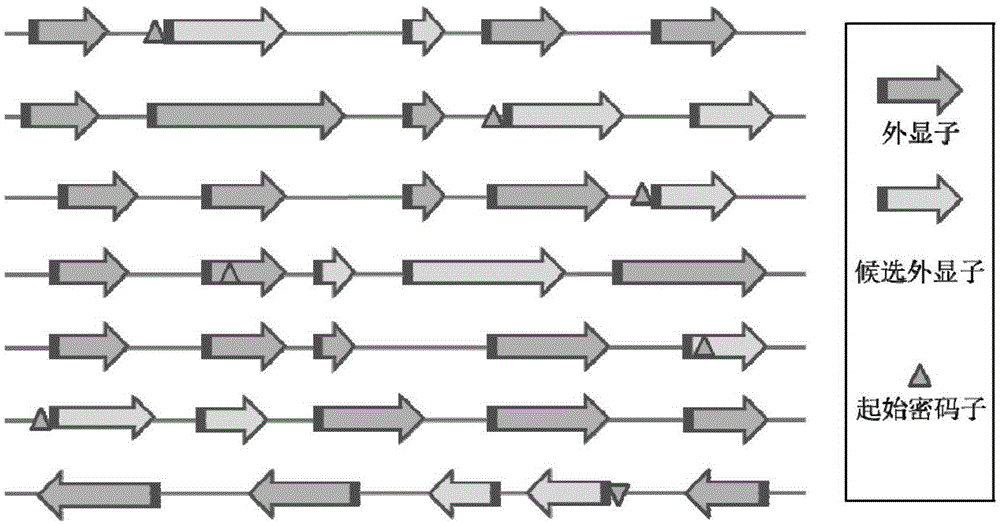
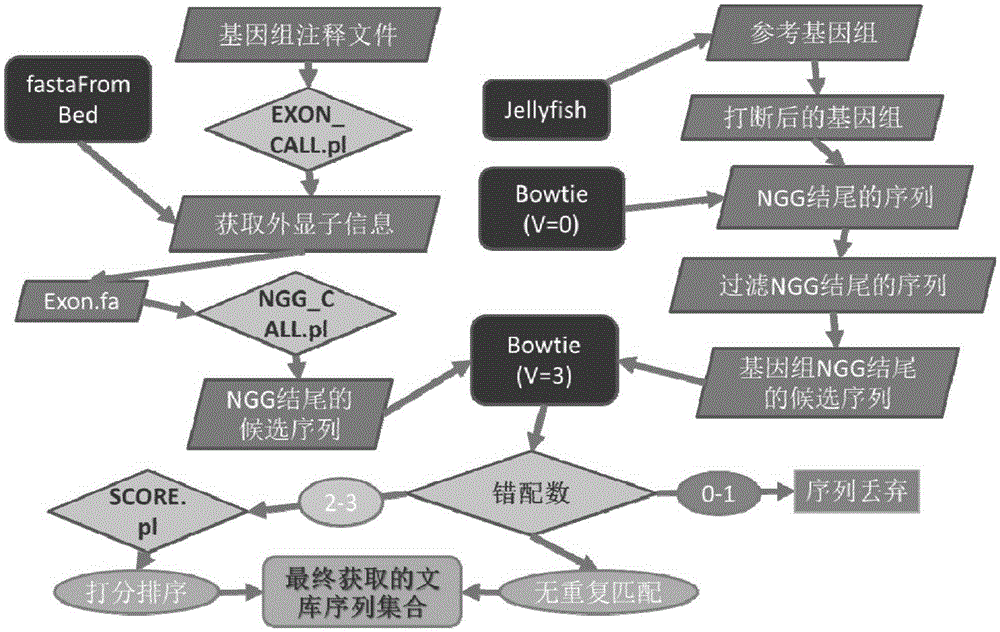
[0061] First, download the chicken reference genome (version number Galgal4, GCA_000002315.2) and its corresponding gene annotation file from the Ensembl database (http: / / www.ensembl.org / index.html). Use the whole genome sequence and gene annotation information to obtain the candidate target 5'-(N of all genes in the genome 20 ) NGG-3' sequence (N stands for A / T / C / G). According to statistics, a total of 380,459 candidate target sequences were obtained in chicken, covering 16,821 genes and 28,915 exons. Then the genome is broken into 23bp fragments and the sequences ending with NGG and without repeats on the genome are screened, compared with the candidate target sequences on the exons, and the corresponding sequences are compared accordi...
Embodiment 2
[0062] Example 2 Screening method for the sgRNA target of the CRISPR-Cas9 system designed for pigs
[0063] In this example, pigs, the representative animal of mammals, were taken as an example to design a genome-wide Cas9 target library.
[0064] First, download the pig reference genome (version number Sscrofa10.2, GCA_000003025.4) and its corresponding gene annotation file from the Ensembl database (http: / / www.ensembl.org / index.html). Use the whole genome sequence and gene annotation information to obtain the candidate target 5'-(N of all genes in the genome 20 ) NGG-3' sequence (N stands for A / T / C / G), statistics show that a total of 626,236 candidate target sequences in pigs were obtained, covering 24,734 genes and 43,049 exons. Then the genome is broken into 23bp fragments and the sequences ending with NGG and without repeats on the genome are screened, compared with the candidate target sequences on the exons, and the corresponding sequences are compared according to the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com