Breast cancer susceptibility gene BRCA1 and BRCA2 detection kit and method
A technology for detecting kits and susceptibility genes, which is applied in biochemical equipment and methods, microbiological determination/inspection, etc., and can solve the problems of low detection throughput, high single-sample detection cost, and long cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0105] The method and kit of the present invention have been used to detect the experimental results of BRCA gene mutations in 313 breast cancer patient tissues, 128 breast cancer patient peripheral blood samples and 46 ovarian cancer patient peripheral blood samples. The experimental results showed that 23 cases of BRCA harmful mutations were detected in 441 breast cancer samples, and 16 cases of BRCA harmful mutations were detected in 46 cases of ovarian cancer samples, and the mutation results were verified by Sanger with 100% accuracy.
Embodiment 2
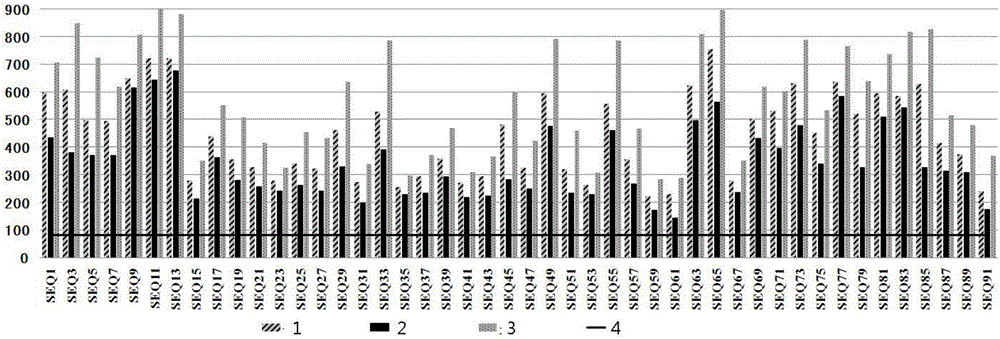
[0107]MiSeq sequencing software splits the sequencing data into corresponding samples according to the provided Index tag sequence information. Then, use BWA software, an alignment program commonly used in the sequencing field, to compare the sequencing sequence of each sample to the human reference genome, and perform the coverage of the target region (that is, the entire coding sequence of BRCA1 and BRCA2) according to the sequencing sequence of the compared genome and multiplex PCR primer uniformity evaluation, the results are as follows figure 2 and 3 shown. (1-average value, 2-minimum value, 3-maximum value, 4-1 / 5 average sequencing depth) The average read volume of 96 samples is between 45,000 and 61,000, the average amplicon depth is 430X, and the target region The coverage rate is 100%, and the coverage rate of the amplicon sequencing depth not lower than 1 / 5 of the average sequencing depth (86X) is 100%. In addition, RT-PCR quantitative analysis of each amplicon f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com