A Virtual Screening Approach for Drug Targets Based on Interaction Fingerprints and Machine Learning
A virtual screening and machine learning technology, applied in instrumentation, informatics, biostatistics, etc., can solve problems such as high false positives and untargeted specific proteins, and achieve the effect of avoiding insufficient fitting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
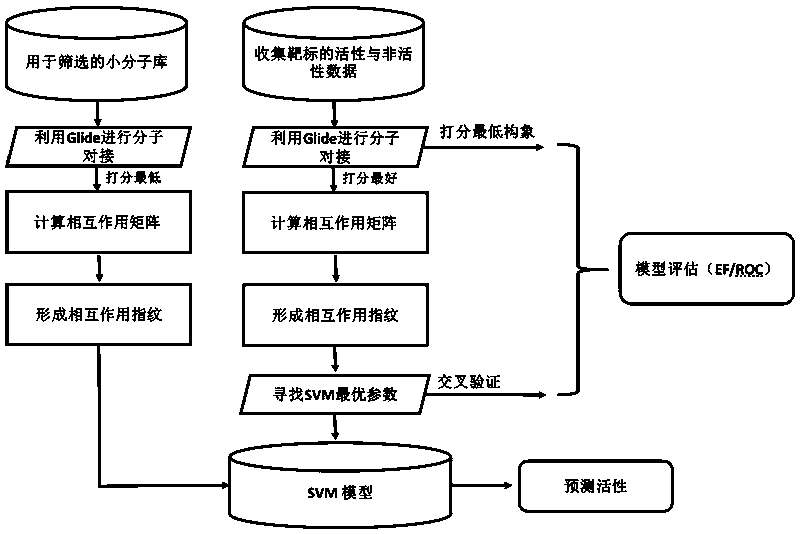
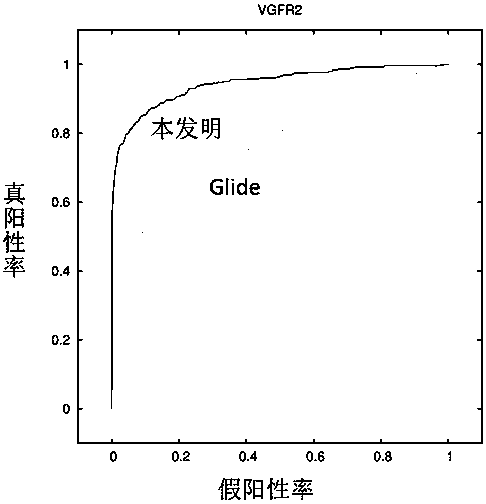
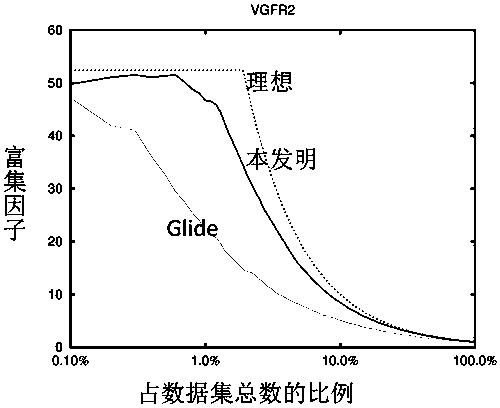
[0073] The present invention will be described in detail by taking the establishment of a screening model of VGFR2 target as an example in conjunction with the accompanying drawings.
[0074] refer to figure 1 , the first thing to do is to change the evaluation index in the SVM software libsvm. Download eval.cpp and eval.h from the libsvm official website, recompile, and change the evaluation criteria of grid search and cross-validation to AUC.
[0075] (1) The activity data of VGFR2 were collected from the DUD-E library, which contained 409 active small molecules and 24950 inactive small molecules. PDB files are 2P2I.
[0076] (2) Calculate the center coordinates of the self-ligand in 2P2I, (38, 35, 12).
[0077] (3) Molecular docking was performed using the Schrödinger molecular docking software Glide.
[0078] (4) Each molecule after docking only adopts the conformation with the lowest GlideScore score. Use the glide_ensemble_merge and glide_sort tools for this purpose...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com