Method for economically and rapidly extracting microbial genome DNA in fermented grains
A microbial and genomic technology, applied in the field of winemaking, to achieve the effect of low labor intensity, saving time and cost, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
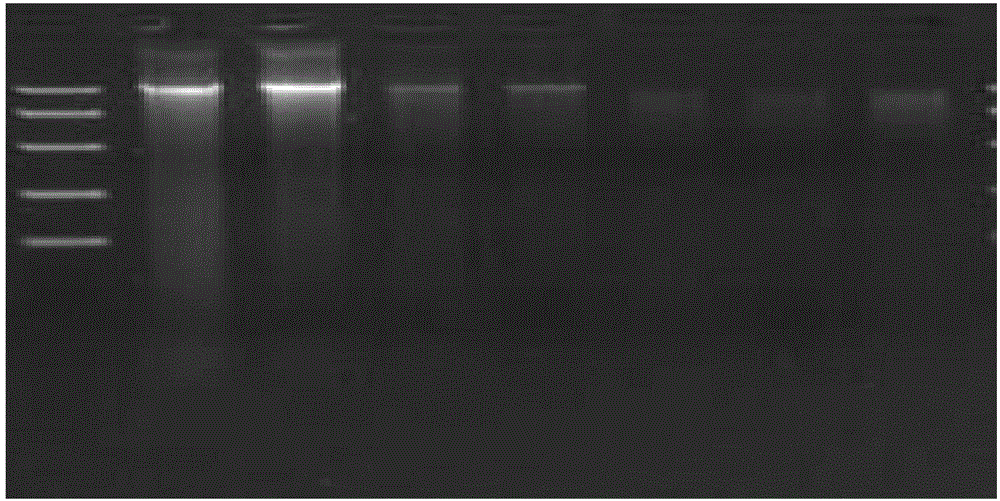
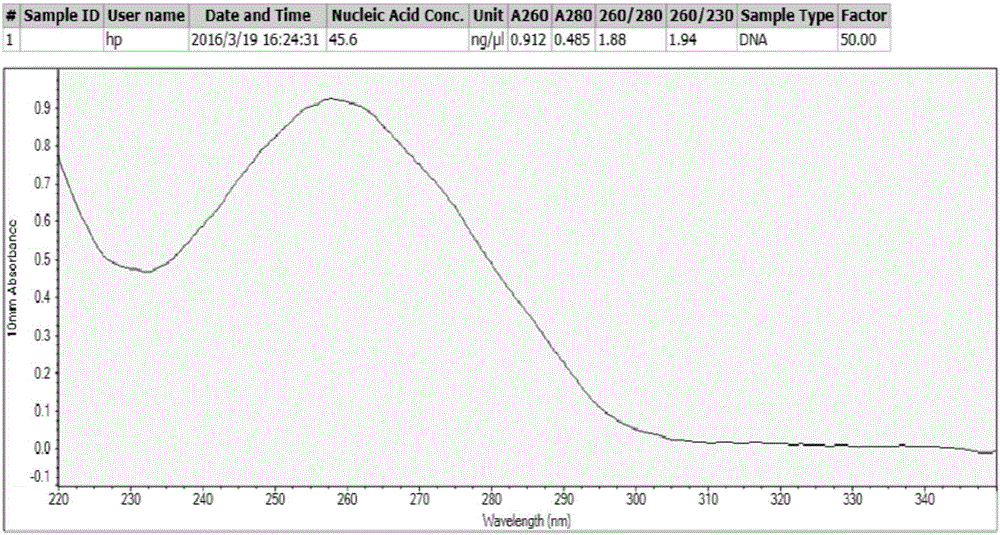
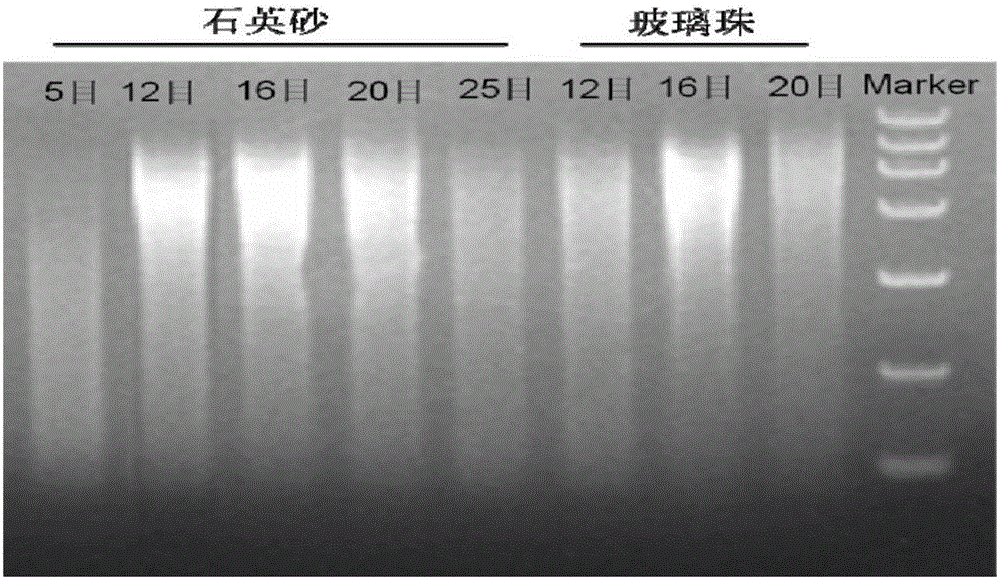
[0070] Example 1 Extraction of Microorganism Genome DNA from Fermented Grains
[0071] 1. After sterilizing and drying the 16-mesh quartz sand, weigh 18.5g and put it into a 50mL sterile centrifuge tube.
[0072] 2. Weigh 7.5g of fermented grains in a centrifuge tube, add 20mL of CTAB extract, oscillate with a homogenizer for 1min, and centrifuge at 10,000rpm for 10min. .
[0073] 3. Transfer the supernatant to a 50mL sterile centrifuge tube, add 0.8-1.2 times the volume of P:C:I (phenol:chloroform:isoamyl alcohol=25:24:1), shake well, and centrifuge at 11000rpm for 20min.
[0074] 4. Transfer the supernatant to a 50mL sterile centrifuge tube, add 1.5-2.5 times the volume of silica gel membrane binding solution, shake well; filter with a 15mL adsorption column, discard the filtrate until the end of the filter.
[0075] 5. Add 3mL of 70% ethanol to the adsorption column, filter and discard the filtrate.
[0076] 6. Repeat step 5 once.
[0077] 7. Centrifuge the empty tube a...
Embodiment 2
[0079] Example 2 Extraction of Microorganism Genome DNA from Fermented Grains
[0080] 1. After sterilizing and drying the 12-mesh quartz sand, weigh 15g and put it into a 50mL sterile centrifuge tube.
[0081] 2. Weigh 6g of fermented grains sample into a centrifuge tube, add 16mL of CTAB extract, oscillate with a homogenizer for 1min, and centrifuge at 10,000rpm for 10min. .
[0082] 3. Transfer the supernatant to a 50mL sterile centrifuge tube, add an equal volume of P:C:I (phenol:chloroform:isoamyl alcohol=25:24:1), shake well, and centrifuge at 11000rpm for 20min.
[0083] 4. Transfer the supernatant to a 50mL sterile centrifuge tube, add 2 times the volume of silica gel membrane binding solution, shake well; filter with a 15mL adsorption column, discard the filtrate until the end of the filter.
[0084] 5. Add 2.5mL 70% ethanol to the adsorption column, filter and discard the filtrate.
[0085] 6. Repeat step 5 once.
[0086] 7. Centrifuge the empty tube at 10000rpm ...
Embodiment 3
[0088] Example 3 Extraction of Microorganism Genome DNA from Fermented Grains
[0089] 1. After sterilizing and drying 20-mesh quartz sand, weigh 22g and put it into a 50mL sterile centrifuge tube.
[0090] 2. Weigh 9g of fermented grains sample into a centrifuge tube, add 24mL of CTAB extract, shake with a homogenizer for 1min, and centrifuge at 10,000rpm for 10min. .
[0091] 3. Transfer the supernatant to a 50mL sterile centrifuge tube, add an equal volume of P:C:I (phenol:chloroform:isoamyl alcohol=25:24:1), shake well, and centrifuge at 11000rpm for 20min.
[0092] 4. Transfer the supernatant to a 50mL sterile centrifuge tube, add 2 times the volume of silica gel membrane binding solution, shake well; filter with a 15mL adsorption column, discard the filtrate until the end of the filter.
[0093] 5. Add 3.5mL 70% ethanol to the adsorption column, filter and discard the filtrate.
[0094] 6. Repeat step 5 once.
[0095] 7. Centrifuge the empty tube at 10000rpm for 2min...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size (mesh) | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com