Application of small-molecule covalent inhibitor in preparing medicine for inhibiting S-adenosylmethionine decarboxylase and screening method of small-molecule covalent inhibitor
A technology of adenosylmethionine decarboxylase and covalent inhibition, applied in the field of biomedicine, can solve problems such as deviation of covalent groups, and achieve the effects of accelerated screening, simple operation and good prediction effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1) Based on the crystal structure of S-adenosylmethionine decarboxylase (AdoMetDC), the Pyr68 residue in the crystal structure of AdoMetDC with PDB ID 3DZ5 was deleted and optimized in Rosetta to obtain the optimized SCAR protein Crystal structure;
[0042] 2) The small molecule compound library used for docking comes from the docking database with ZINC (http: / / zinc.docking.org). The search conditions for small molecules are: ①SMILES expression: CONN, and select the substructure of "substructure" to search Molecular structure containing the above groups; ② molecular weight greater than 200 and less than 400; ③ xLogP ≤ 3.5; ④ rotatable bond greater than or equal to 3 and less than or equal to 9; ⑤ hydrogen bond donor greater than or equal to 2 and less than or equal to 10; ⑥ hydrogen bond The receptor is greater than or equal to 2 and less than or equal to 10. After the server returns the search results, the structure of the corresponding small molecule is obtained;
[...
Embodiment 2
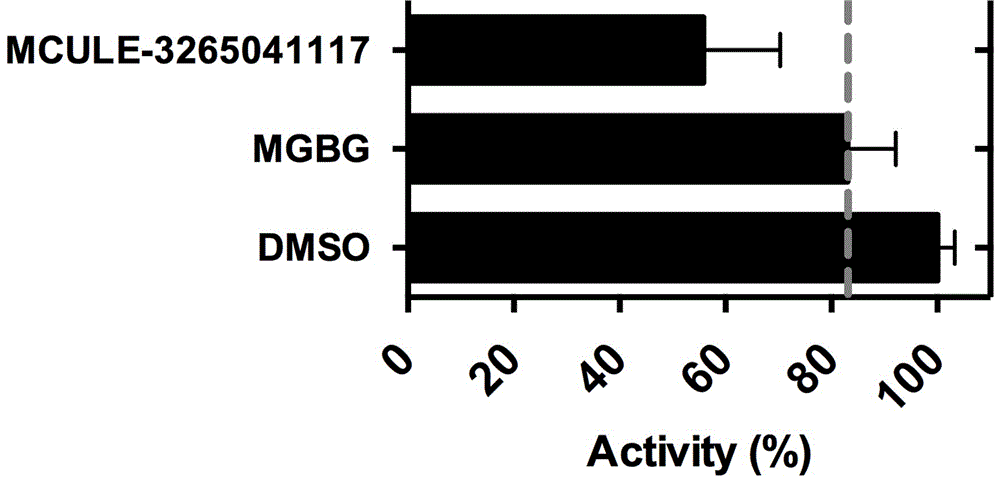
[0047] Utilizing the method of the patent (ZL201410530672.3), we have carried out AdoMetDC activity evaluation, spectroscopic detection described step 4) the activity inhibitory effect of the small molecule covalent inhibitor of Example 1 on AdoMetDC, small molecules mixed with AdoMetDC in After co-incubating at 37 degrees for 30 minutes, add the substrate and react for 5 minutes before testing. DMSO is used as the solvent control, and AdoMetDC inhibitor MGBG (mitoguanidine hydrazone) is used as the positive control. The results are shown in figure 1 , this small molecule covalent inhibitor has better inhibitory ability than MGBG;
Embodiment 3
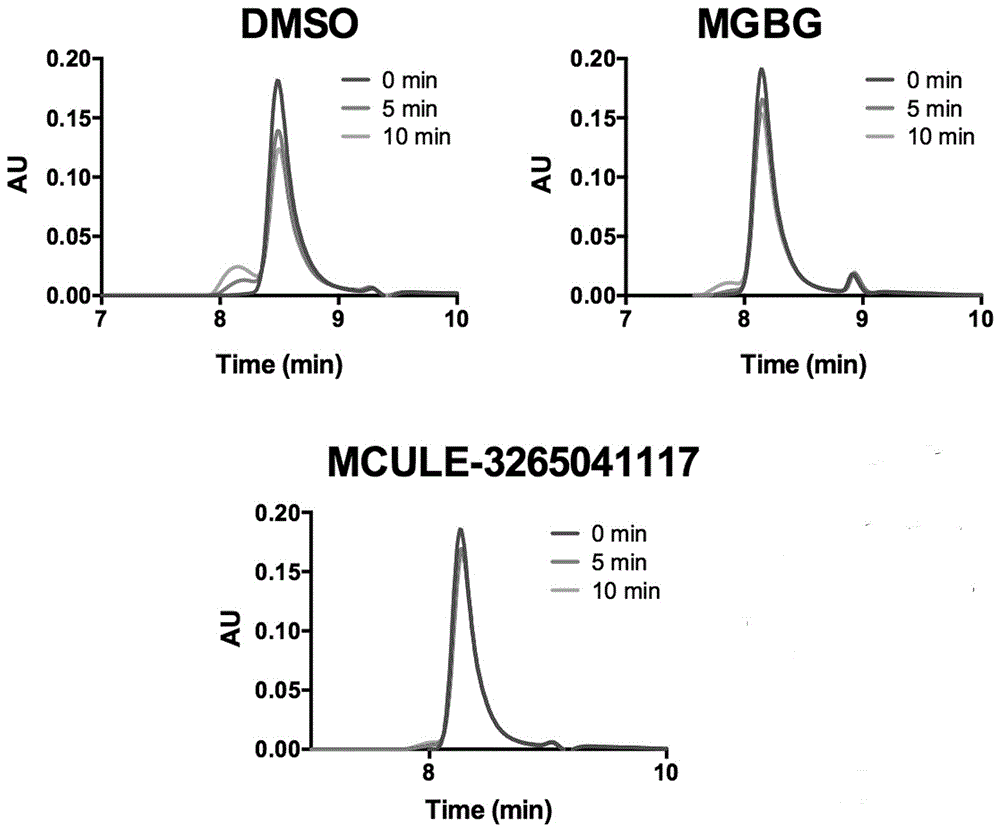
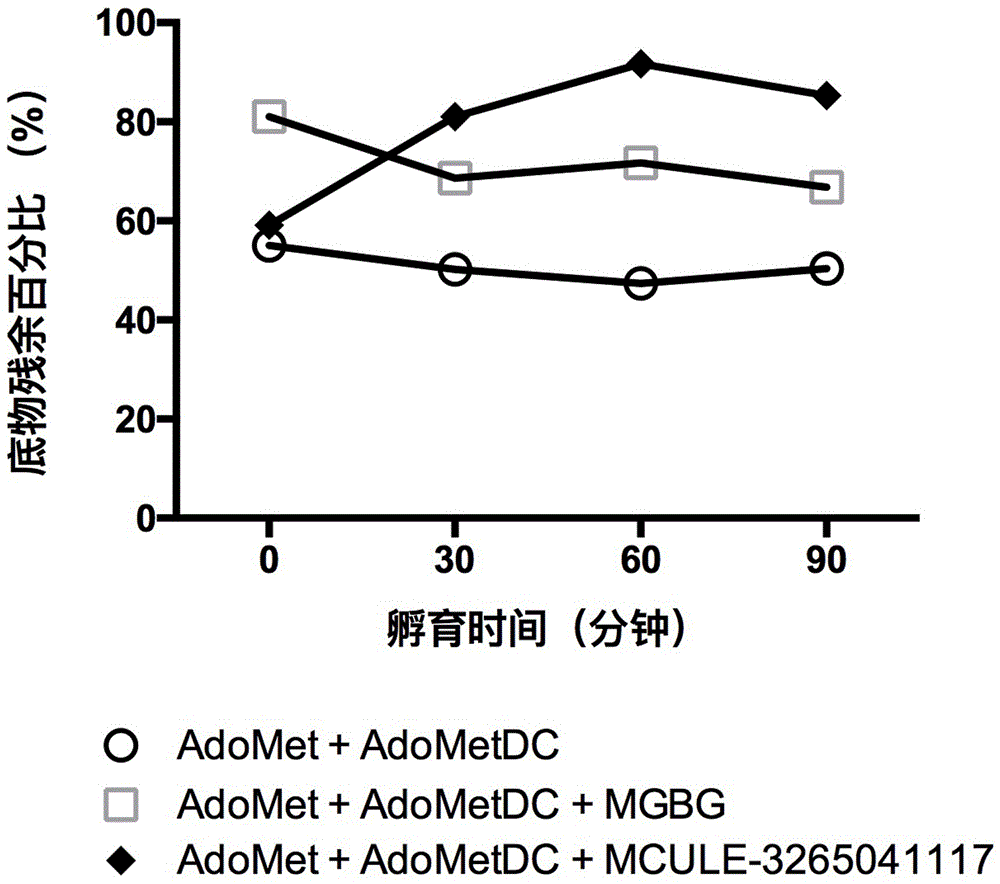
[0049] The inhibitory effect of the small molecule in Example 1 on AdoMetDC was verified by detecting the consumption of the substrate AdoMet by using high performance liquid chromatography (HPLC). The instrument used for HPLC is Waters e2695, the column is Waters C18 reverse phase column (5um, 4.6x250mm), the column temperature is 30 degrees, the mobile phase is 0.01mol / L ammonium formate (pH 3.5):methanol=97:3, and the flow rate is 0.5mL / min , detection wavelength 254nm. Before loading the sample, add 9 times the volume of methanol to stop the reaction, centrifuge at 12000g for 10 minutes to remove the denatured protein, then take the supernatant sample for analysis, the results are shown in figure 2 ,Depend on figure 2 It can be seen that after mixing and incubating the small molecule and AdoMetDC at 37 degrees for 30 minutes, the reaction was terminated after adding the substrate for 0 minutes, 5 minutes or 10 minutes, and the consumption of the substrate was detected. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com