Local Lipschitz support surface-based dual-layer differential evolution protein structure prediction method
A protein structure and prediction method technology, applied in the field of double-layer differential evolution protein structure prediction, can solve the problems of inability to accurately search for the lowest energy conformation, time-consuming, expensive and labor-intensive, and high purity requirements.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] The present invention will be further described below in conjunction with the accompanying drawings.
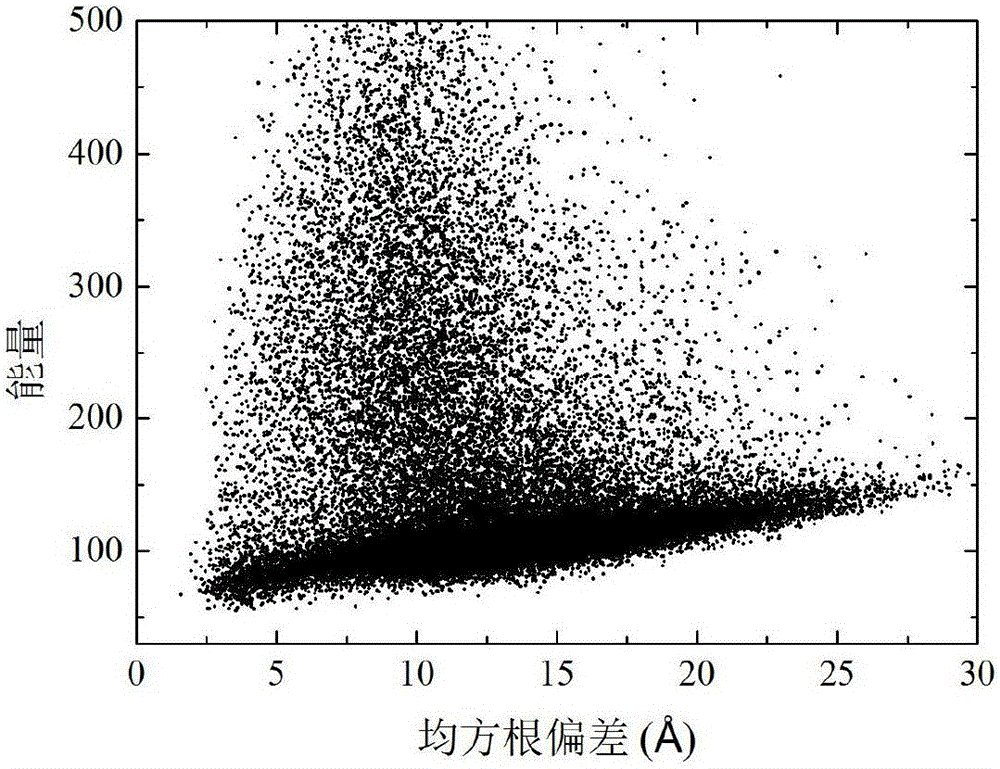
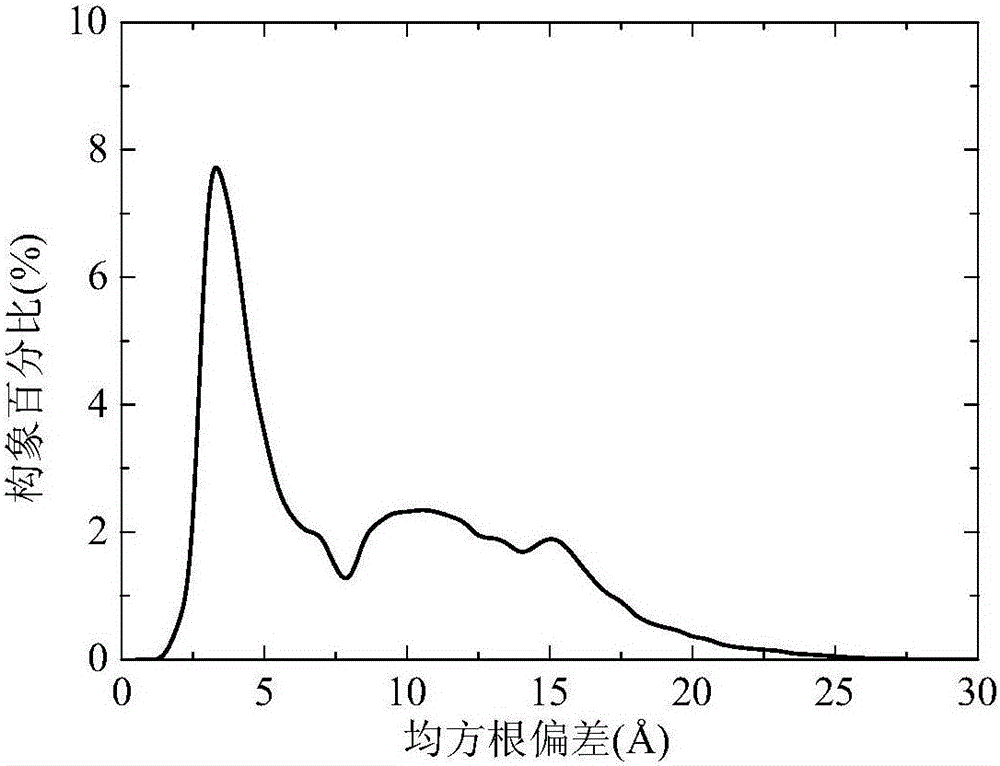
[0038] refer to Figure 1-3 , a two-layer differential evolution protein structure prediction method based on local Lipschitz support surfaces, including the following steps:
[0039] 1) Select the force field model:
[0040] The expression of the energy function using the Rosetta force field model is as follows
[0041] E = W int e r r e p E int e r r e p ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com