Cascade amplifying-strategy biomolecule detecting method based on self-locking aptamer probe
A self-locking technology of small biomolecules, applied in the field of high sensitivity and high specificity detection, it can solve problems such as interference signal, double-strand structure blocking chain leakage, etc., to achieve high sensitivity and high specificity detection, large application Potential, the effect of achieving high sensitivity and high specificity detection
Active Publication Date: 2016-06-01
SHANDONG UNIV
View PDF3 Cites 17 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
However, due to the short length of the blocking strand (12-15nt), the instability of the double-stranded structure will cause the blocking strand to leak (fall off from the aptamer probe), which in turn leads to the generation of interfering signals
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
[0058] 1. Experimental part
[0059] 1.1 Reagents and materials
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
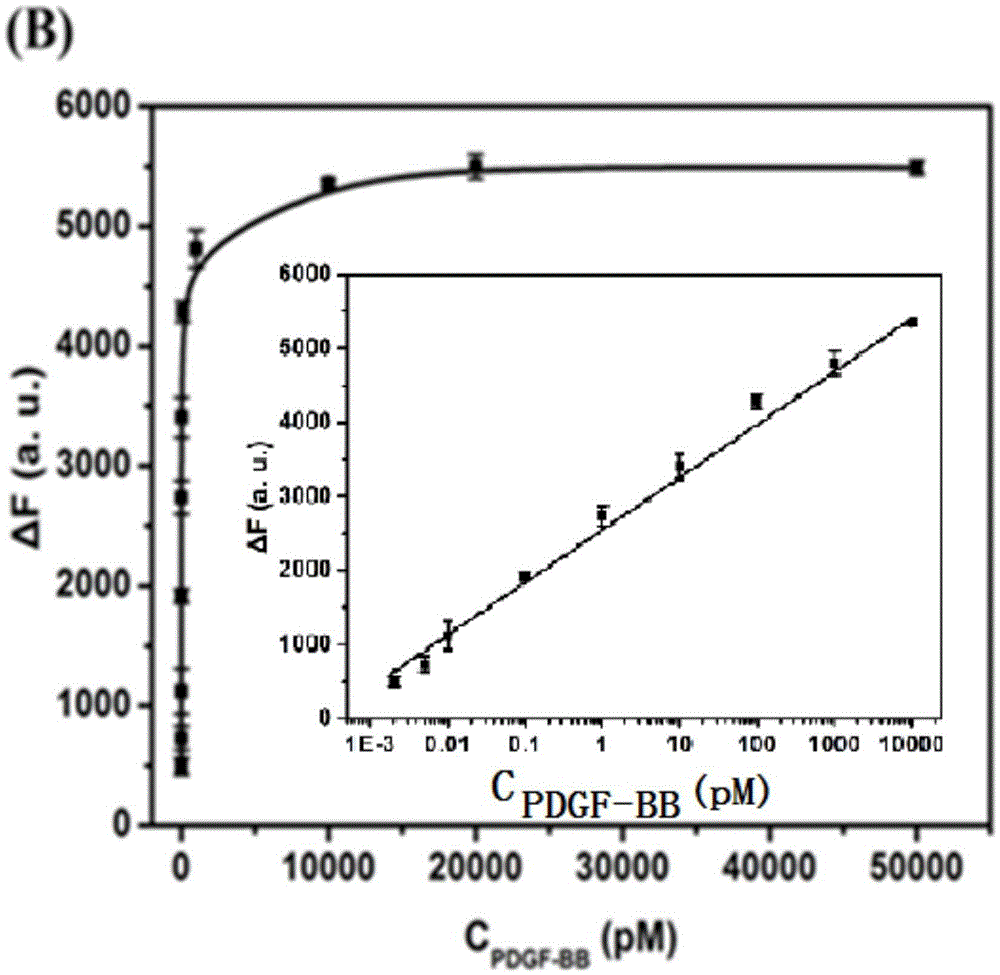
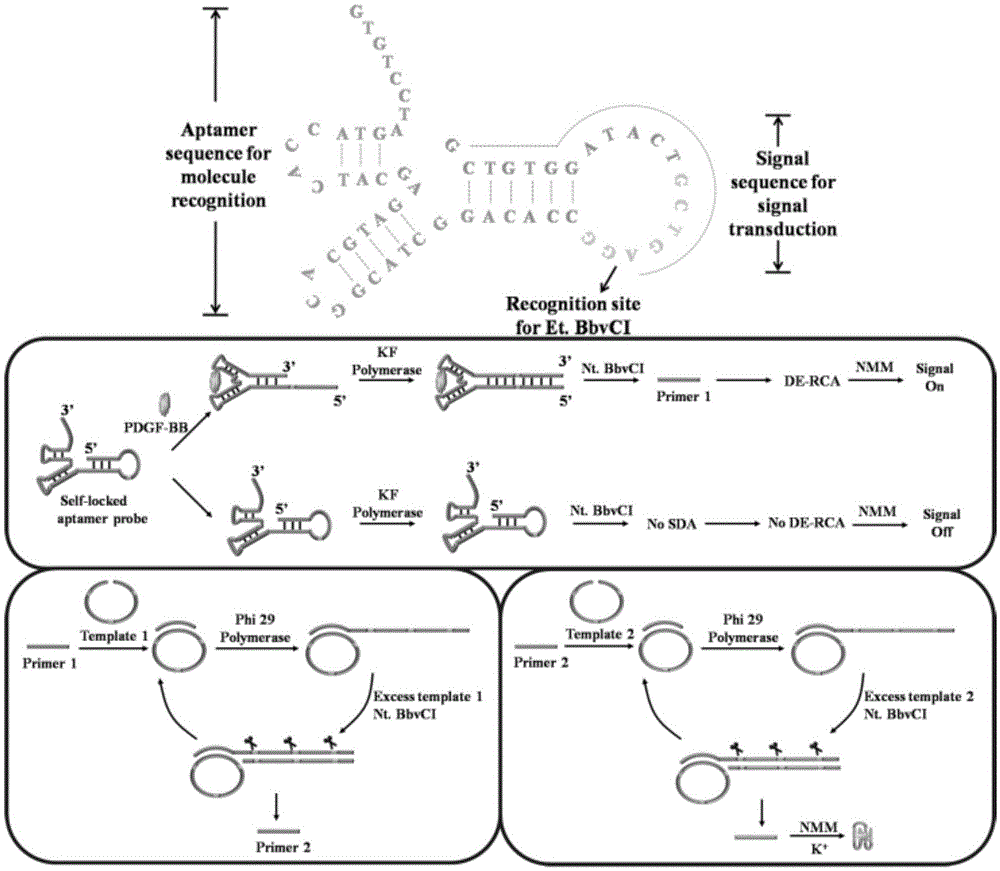
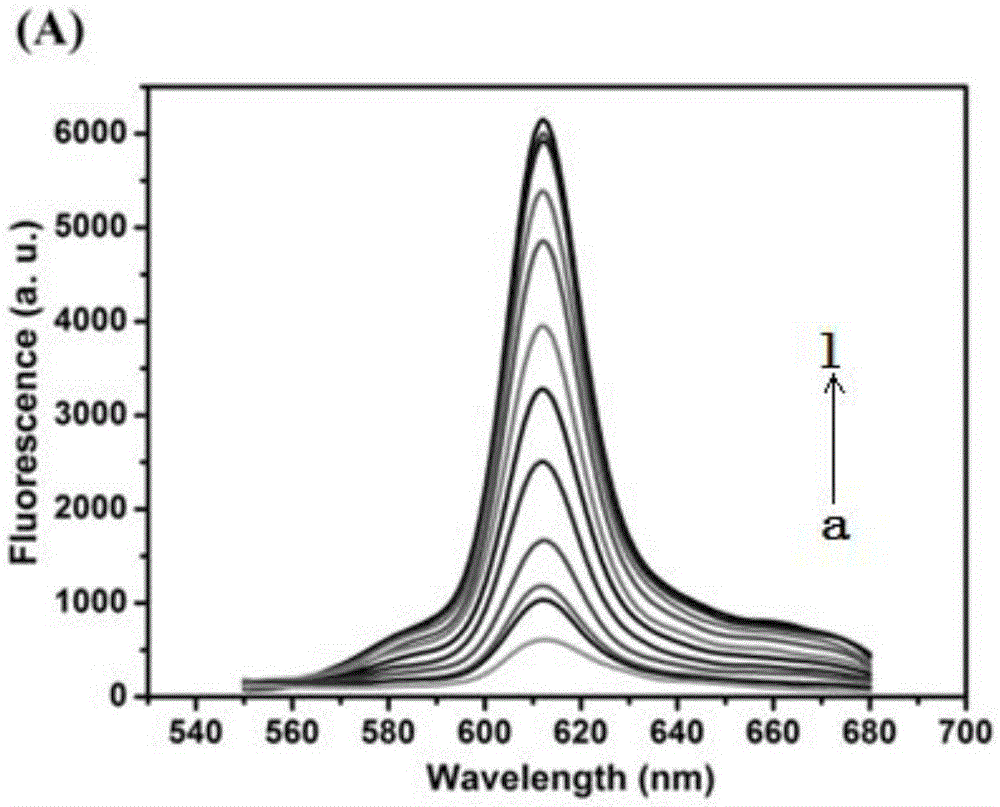
The invention discloses a cascade amplifying-strategy biomolecule detecting method based on a self-locking aptamer probe. The probe at least comprises two parts of an aptamer sequence of a 3(minute) terminal having specific recognition on a target and a signal transduction sequence of a 5(minute) terminal, wherein the signal transduction sequence of the 5(minute) terminal is hybridized with a part of the aptamer sequence to form a stem-loop structure of the 5(minute) terminal, and the signal transduction sequence of the 5(minute) terminal comprises a recognition site of Cech endonuclease. The probe combines strand displacement amplification (SDA) with a double-exponential rolling circle amplification (DE-RCA) strategy to achieve ultra-sensitive and high-specific detection on protein-PDGF-BB and micromolecule-adenosine, wherein the detection limit reaches 3.8*10<-16>mol / L and 4.8*10<-8>mol / L respectively.
Description
technical field [0001] The invention relates to the technical field of nucleic acid detection, in particular to a highly sensitive and specific detection method for proteins and small molecular substances using a self-locking aptamer probe-based cascade amplification strategy. Background technique [0002] Aptamers are a class of artificially synthesized, short single-stranded oligonucleotide sequences that are often used in biosensing, diagnosis, and therapy. Aptamers usually have specific and compact secondary or tertiary structures, and have high affinity and selectivity for their corresponding targets. Compared with antibodies or antibody fragments, aptamers are stable, easy to synthesize and modify, and have a wide range of targets, including proteins, small molecules, metal ions, and even cells. Therefore, aptamers have been widely used in the construction of molecular probes. [0003] Aptamer-based molecular probes mainly include two parts: aptamer sequence and sign...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Applications(China)
IPC IPC(8): C12N15/115G01N33/68
CPCC12N15/115C12N2310/16C12N2310/531C12Q2531/119C12Q2531/125C12Q2563/107G01N33/68
Inventor 王磊姜玮李伟
Owner SHANDONG UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com