Methods and kits for the molecular subtyping of tumors
A technology for molecular subtypes and tumors, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve problems such as technical differences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0320] Example 1: Determination of mRNA expression levels by reverse transcription (RT) quantitative PCR (RT-qPCR)
[0321] RNA was isolated from paraffin-embedded formalin-fixed tissues (=FFPE tissues). More specifically, total RNA was extracted from 5–10 μm rolled FFPE tumor tissues using the High Purity RNA Paraffin Kit (Roche, Basel, Switzerland) or XTRAKT RNA Extraction Kit XL (Stratifyer Molecular Pathology, Cologne, Germany), and quantified by Ribogreen RNA (MolecularProbes, Eugene, OR) to quantify and identify the quality by real-time fluorescent RT-PCR of the reference gene RPL37A fragment. It was found that there were differences between different extraction methods when comparing the quantitative data of target genes in serial sections by different methods. For the purposes of the present invention, it is preferred to use the XTRAK T RNA Extraction Kit XL. 2.5 μl of each qualifying RNA extract (approximately 50-100 ng) was assayed by qRT-PCR as described below. ...
Embodiment 2
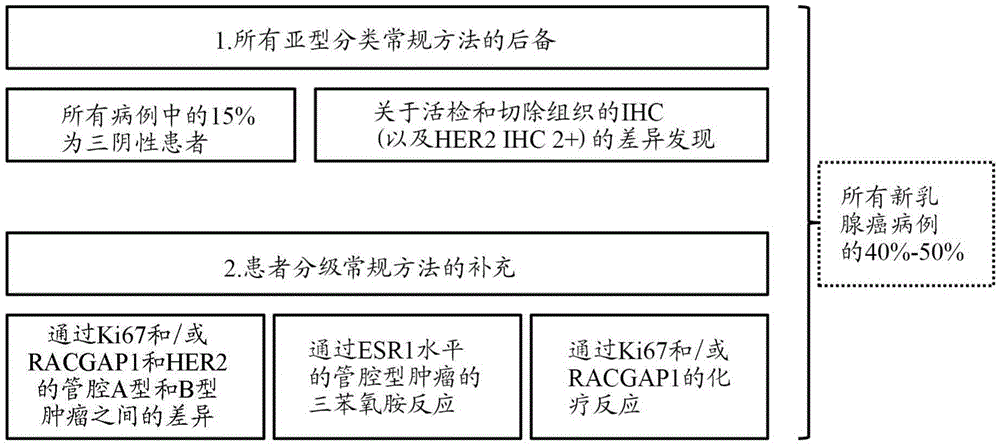
[0326] Example 2: mRNAs based on HER2, ESR1, PGR, Ki67, and optionally RACGAP1 Molecular subtyping of tumors by expression levels
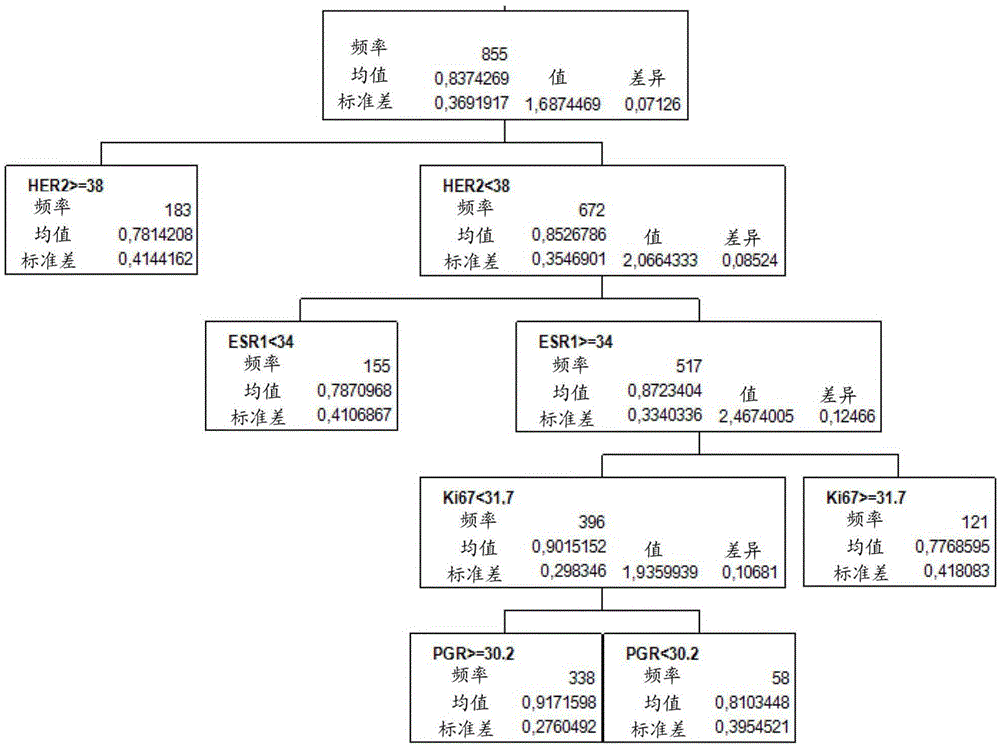
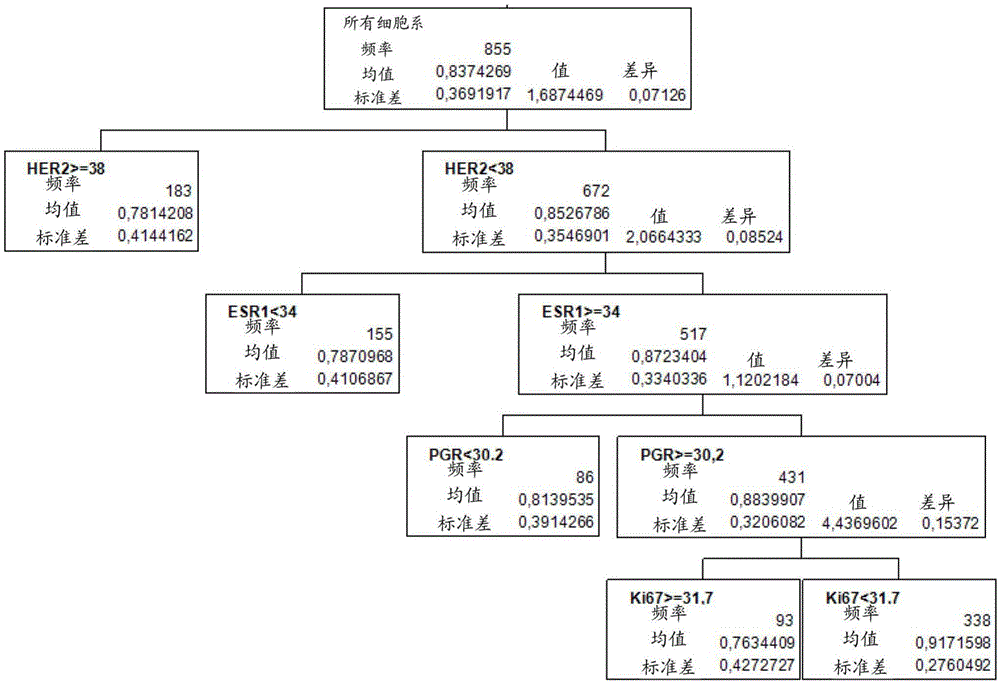
[0327] For the assessment of breast tumors, 855 out of 1010 breast cancer patients from the FinHER study (Joensuu et al. (2006), NEngl J Med, 354:809-820) can be used. The average threshold values (given as 40-ΔCT values) are as follows: HER2: 38; ESR1: 34; PGR: 30,2; Ki67: 31,7; Based on the evaluation of previous studies (Koutrasetal. (2008), Brit.J.ofCanc., 99: 1775-1785; Pentheroudakis etal. (2009), Breast CancerResTreat2009, 116: 131-143), HER2 and ESR1 were determined using 268 samples. threshold, and applied to the FinHER study (proportional hazards model validation). CALM2 was used as a reference gene.
[0328] Tumors were assigned to molecular subgroups based on the mRNA expression levels of HER2, ESR1, PGR, and Ki67 (negative / low indicating expression levels below the set expression threshold; positive / increasing indicating expre...
Embodiment 3
[0339] Example 3: Measurement of biomarkers HER2, mRNA expression levels of ESR1, PGR, Ki67 and determination of molecular subtypes
[0340] RNA was isolated from FFPE tissue. More specifically, total RNA was extracted from 10 μm FFPE breast tumor tissue sections using the XTRAKT RNA extraction kit (Stratifyer Molecular Pathology, Cologne, Germany). The RNA eluate was used directly without concentration determination. 2.5 μl of RNA from each extract was assayed by RT-qPCR as described below.
[0341] For RT-qPCR, primers flanking the target region and hydrolysis probes 5’-fluorescently labeled with 3’-TAMRA or 3’-Dabcyl quenchers were used for each target. The primers and probes used are listed in Table 1. To correct for varying amounts of sample RNA, the genes CALM2 and B2M were used as reference genes to normalize expression results. RT-qPCR was performed in duplicate with the following combination: HER2 / ESR1, Ki67 / B2M, and PGR / CALM2. Each of the three 4x assay mixes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com