Support vector machine based antiviral inhibitor dissociation rate constant prediction method
A technology of support vector machines and rate constants, applied in the field of bioinformatics, which can solve the problems of drug development loss, no consideration of complex binding kinetics, slow dissociation rate, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Take the influence of the antiviral inhibitor structure on the dissociation rate as an example to specifically illustrate the implementation process of the present invention:
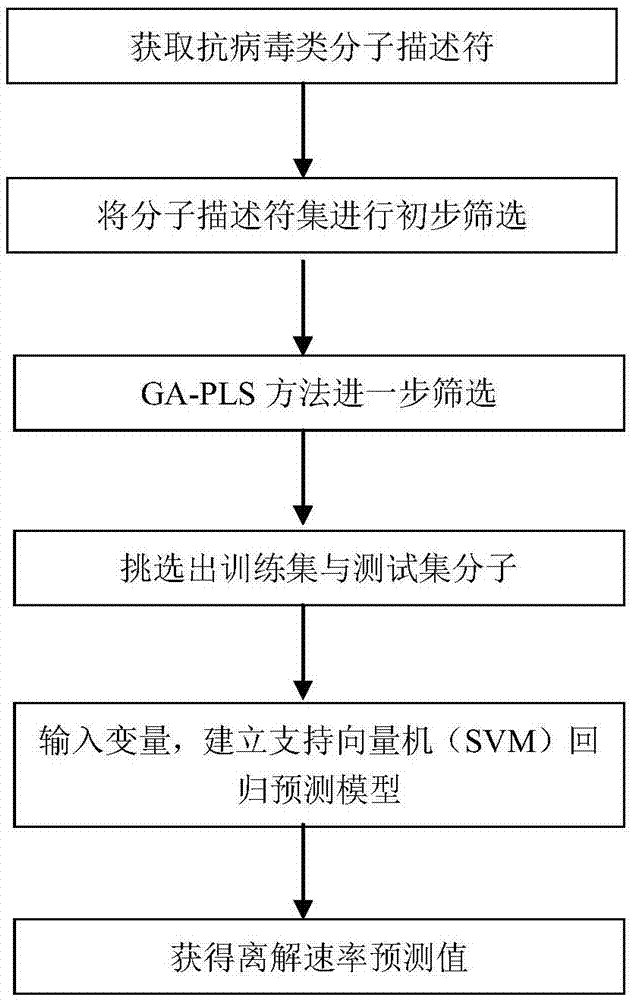
[0054] (1) Acquire molecular descriptors. The molecular descriptor calculation software used in this experiment is Dragon1.1. Input the collected antiviral inhibitor molecules into Dragon, and the calculation software will calculate more than 1600 descriptors in 20 categories. Antiviral Inhibitor K off The value is logarithmically processed, thus establishing a logK off The data sets corresponding to the corresponding inhibitor descriptors are shown in Table 1 (see Table 1 on the attached page).
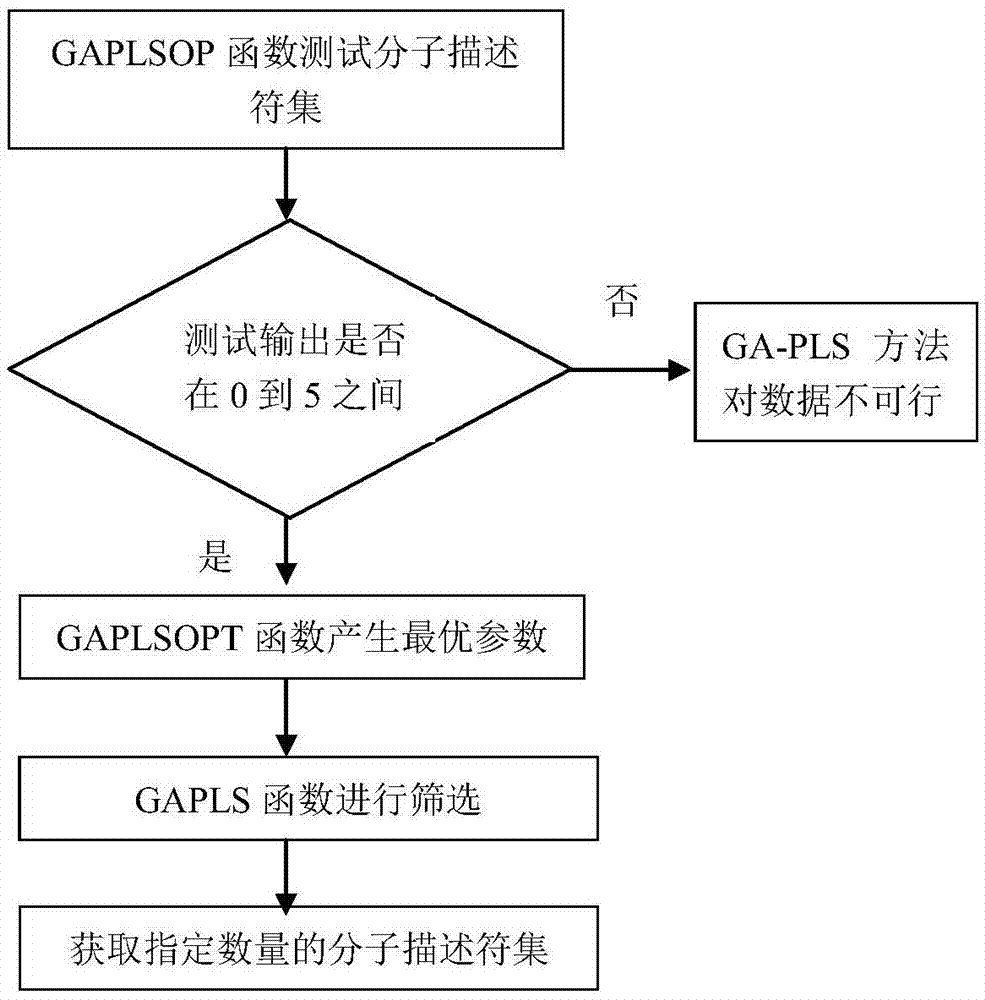
[0055] (2) Preliminary screening of the data set. The preliminary screening tool we choose is SPSS software. Import the data in step (1) into SPSS to conduct preliminary screening on the data set. The specific screening steps are as follows:
[0056] (2-1) Descriptors having the same data in 90% ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com