A method of acquiring an unknown flanking sequence of a known sequence
A technology of unknown sequence and sequence, which is applied in the field of genome unknown sequence amplification, which can solve the problems of low probability of effective amplification, affecting the success rate, and many non-specific amplifications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] 1. DNA extraction and purification:
[0026] Various general DNA extraction methods and purification methods can be used to extract and obtain purified genomic total DNA.
[0027] 2. Primer design:
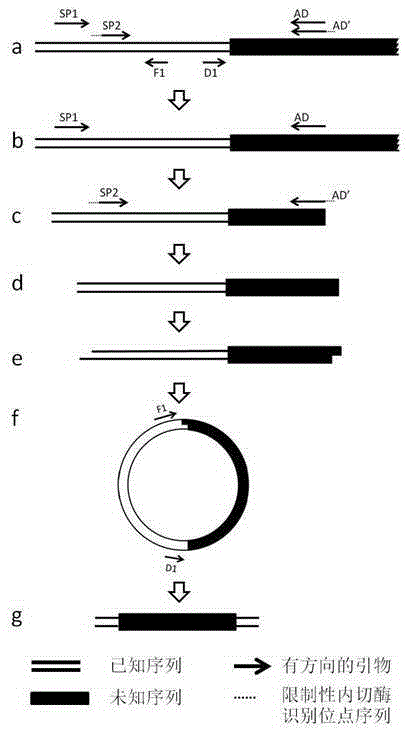
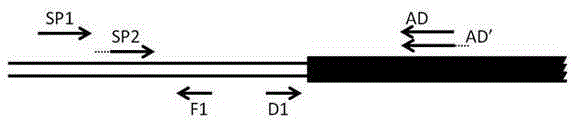
[0028] Select the sequence of the middle fragment of A. clematis PAL gene obtained through homologous amplification, and design two specific primers in the same direction. The sequence from the sequence far away from the boundary sequence to the sequence close to the boundary sequence is marked as SP15'-TGGTGTCACTACTGGATTTGG-3' and SP25 '-CCCGGGGCATCCGATTCGAAATCCT-3', SP2 has a restriction endonuclease recognition site 5'-CCCGGG-3', and a pair of reverse PCR primers F1 is designed between SP2 and the border sequence: 5'-CAAGATCGCCGGAGGCGGTG-3' and D1: 5'-CTTACATTGCGGGTGTCTTAACC-3', as in figure 2 .
[0029] 3. Unknown sequence amplification:
[0030] A. The first amplification with asymmetric primers:
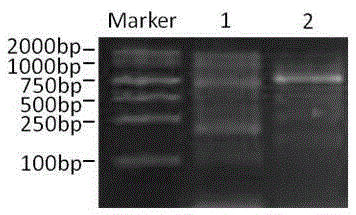
[0031] Using genomic DNA as a template, the first round of PCR am...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com