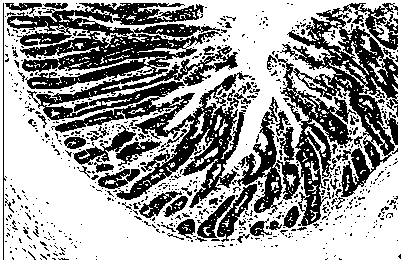
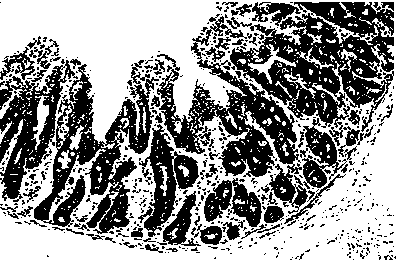
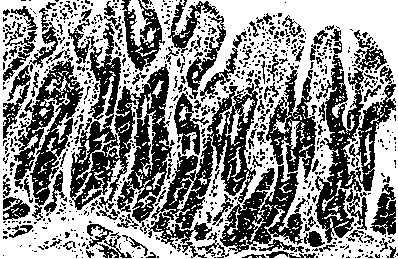
Application of serum ssc-mir-194b as a molecular marker for detecting intestinal stress injury in piglets
A technology of ssc-mir-194b and serum, applied in the field of livestock genetic engineering, can solve problems such as lack of diagnostic value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: In this example, ssc-miR-194b was obtained by analyzing the correlation between serum microRNA and intestinal microRNA of weaned piglets, including the following steps:
[0024] 1. Total RNA extraction from piglet intestinal samples
[0025] Twenty-four piglets (6 piglets per litter) were selected from 4 litters and 25-day-old piglets with the same parity and date of birth, and were divided into lactation group and weaning group (weaning on the same day). The piglets in the weaning group were slaughtered 1d, 4d and 7d after weaning, and 4 piglets (1 per litter) were slaughtered in each group. Jejunal tissue samples were collected, and total RNA samples of piglet intestinal tissue were extracted by Trizol method.
[0026] 2. Small molecule RNA library construction
[0027] The total RNA samples of 4 biological replicates in each treatment group were converted by concentration and mixed in equal amounts to form RNA sample pools, and 15% denaturing polyacrylam...
Embodiment 2
[0051] Example 2: In this example, the ssc-miR-194b obtained in Example 1 was used to mark intestinal stress injury in weaned piglets.
[0052] (1) Randomly select 24 weaned piglets and mark them as Nos. 1 to 24, collect whole blood of weaned piglets, and prepare serum samples of Nos. 1 to 24;
[0053] (2) Using Plasma / Serum Exosome Kit (Norgen, 49200) reagent to extract total RNA in serum samples;
[0054] (3) using miRNeasy Mini Kit (Qiagen 217004) kit to extract serum microRNA in Total RNA;
[0055] (4) Use the ssc-miR-194b reverse transcription primer shown in SEQ ID NO.2 to reverse transcribe the microRNA to obtain cDNA: the reverse transcription system is: microRNA sample 2.5ul (40ng / ul), ssc-miR-194b reverse transcription primer 0.5ul (2uM), dNTP 0.25ul (10mM each),
[0056] 5×RT buffer 1.0ul, RNase inhibitor 0.25ul (40U / ul), M-MLV 0.5ul (200U / ul), the total volume of the reaction system is 5.0ul, put the uniformly mixed reaction system at 42°C for 60min, and then pla...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com