Probe preparation method for multi-gene capture sequencing
A multi-gene and probe technology, applied in biochemical equipment and methods, DNA preparation, microbial measurement/inspection, etc., can solve problems such as expensive, difficult to promote, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
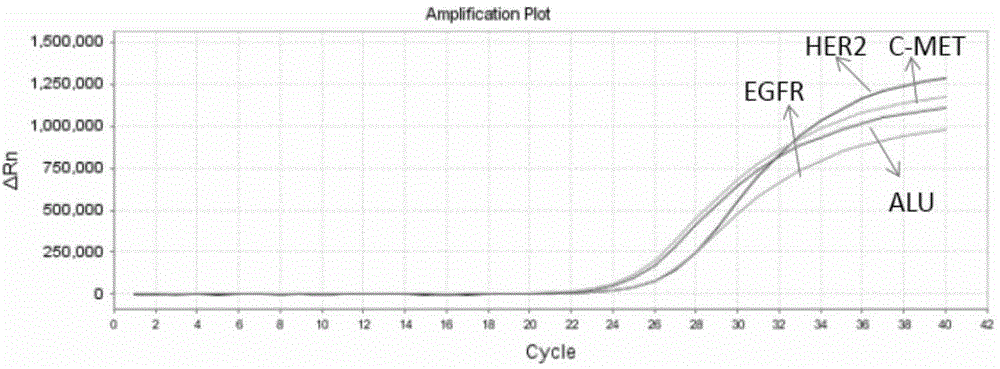
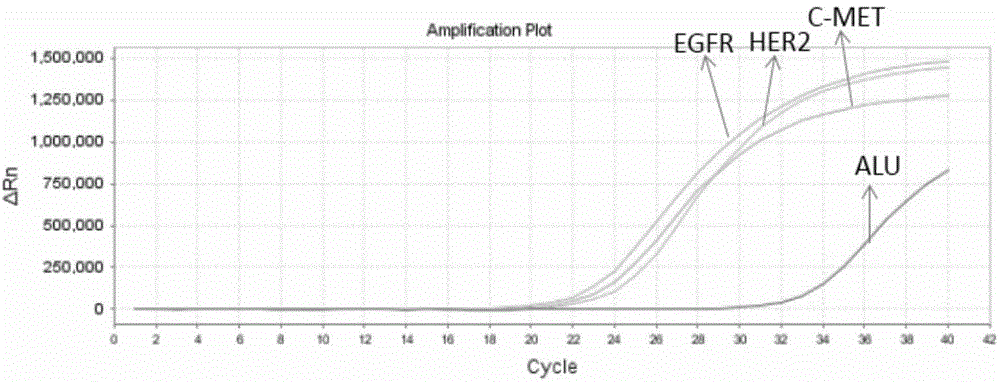
[0061] Please see attached figure 1 and attached figure 2 As shown, a method for preparing probes for multi-gene capture sequencing, the method at least includes the following steps:
[0062] Step 1: Prepare probes.
[0063] Step 2: Capture target DNA.
[0064] Step 3: Carry out the elution procedure.
[0065] Step 4: Use a high-fidelity enzyme reaction amplification system to amplify after capturing the target DNA.
[0066] Step 5: After the amplified product is purified with magnetic beads, it is quantified to 20 ng / ul with Nanodrop, and stored in aliquots.
[0067] Step 6: Use the Real-time PCR method to detect the positive control sequence to verify the capture efficiency.
[0068] In said step 1, at least the following sub-steps are included:
[0069] Step 1.1: Synthesize 12K, 60K, 90K and other chips with different oligonucleotide quantities. The chips contain a 120bp target region and a common 15-base end:
[0070] 5'-ATCGCACCAGCGTGTN120CACTGCGGCTCCTCA-3'.
[0...
Embodiment 1
[0115] Preparation of capture probes targeting cancer-related genes. A mixture of 120bp+30bp fragment lengths of 319 related genes (as shown in Supplementary Table 1) was prepared in total.
[0116] Step 1: Prepare probes.
[0117]Step 1.1: Synthesize chips with 12K, 60K, 90K and other different oligonucleotide quantities, these chips contain 120bp target regions and common 15 base ends.
[0118] 5′-ATCGCACCAGCGTGTN120CACTGCGGCTCCTCA-3′
[0119] Step 1.2: Dissolve the synthesized oligonucleotide library in 400ul low concentration TE (10mM Tris-HCL, pH8, 0.1mM EDTA).
[0120] Step 1.3: Since the concentration of each oligonucleotide is at the fmol level, amplification is required to obtain the desired concentration.
[0121] Primer sequence: A 5′Biotin-CTGGGAATCGCACCAGCGTGT-3′
[0122] B 5′-CGTGGATGAGGAGCCGCAGTG-3′
[0123] Amplification conditions: (prepare three 50ul PCR mixes), templates are 1ul, 2ul, 5ul,
[0124]
[0125]
[0126] The PCR reaction conditions ar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com