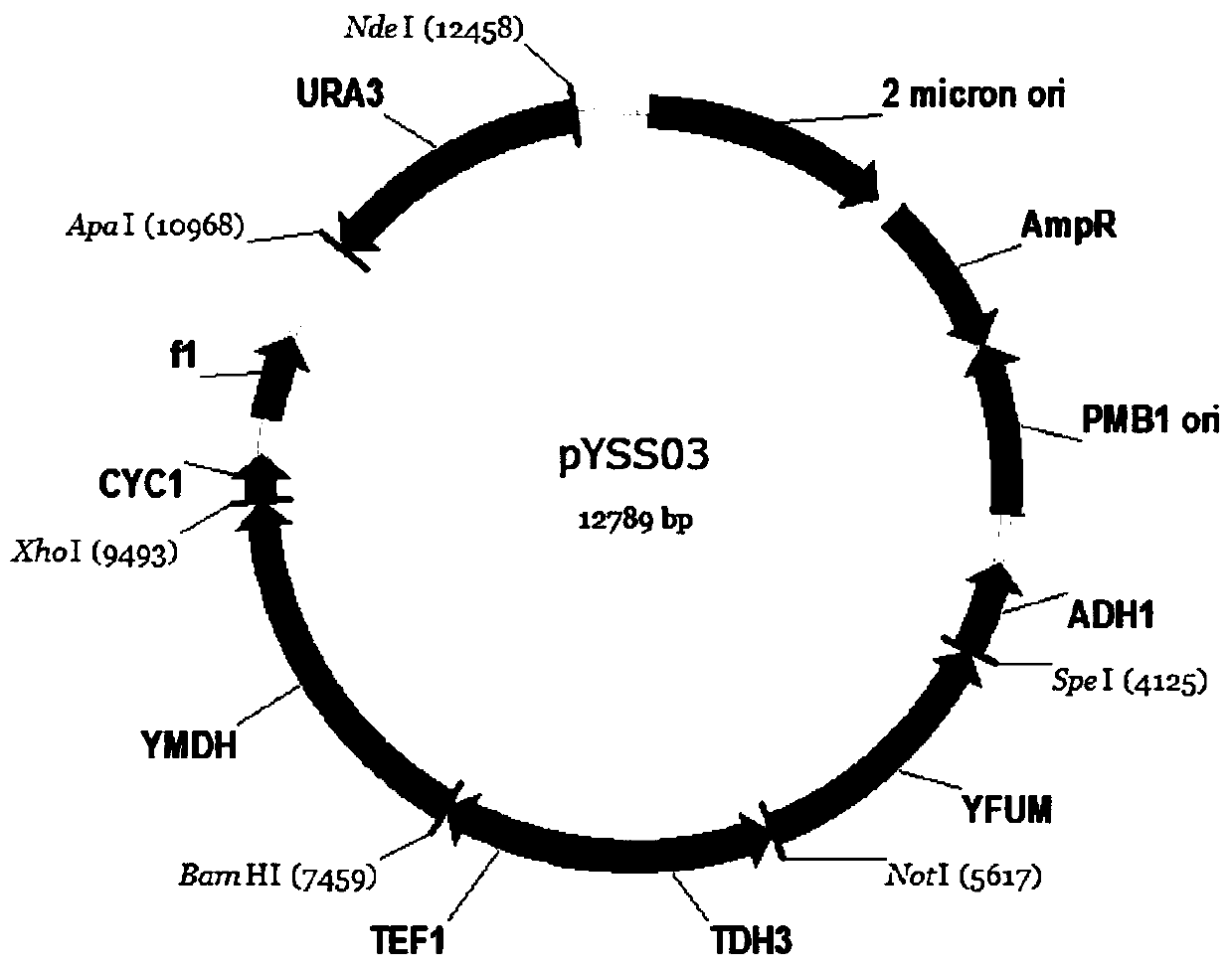
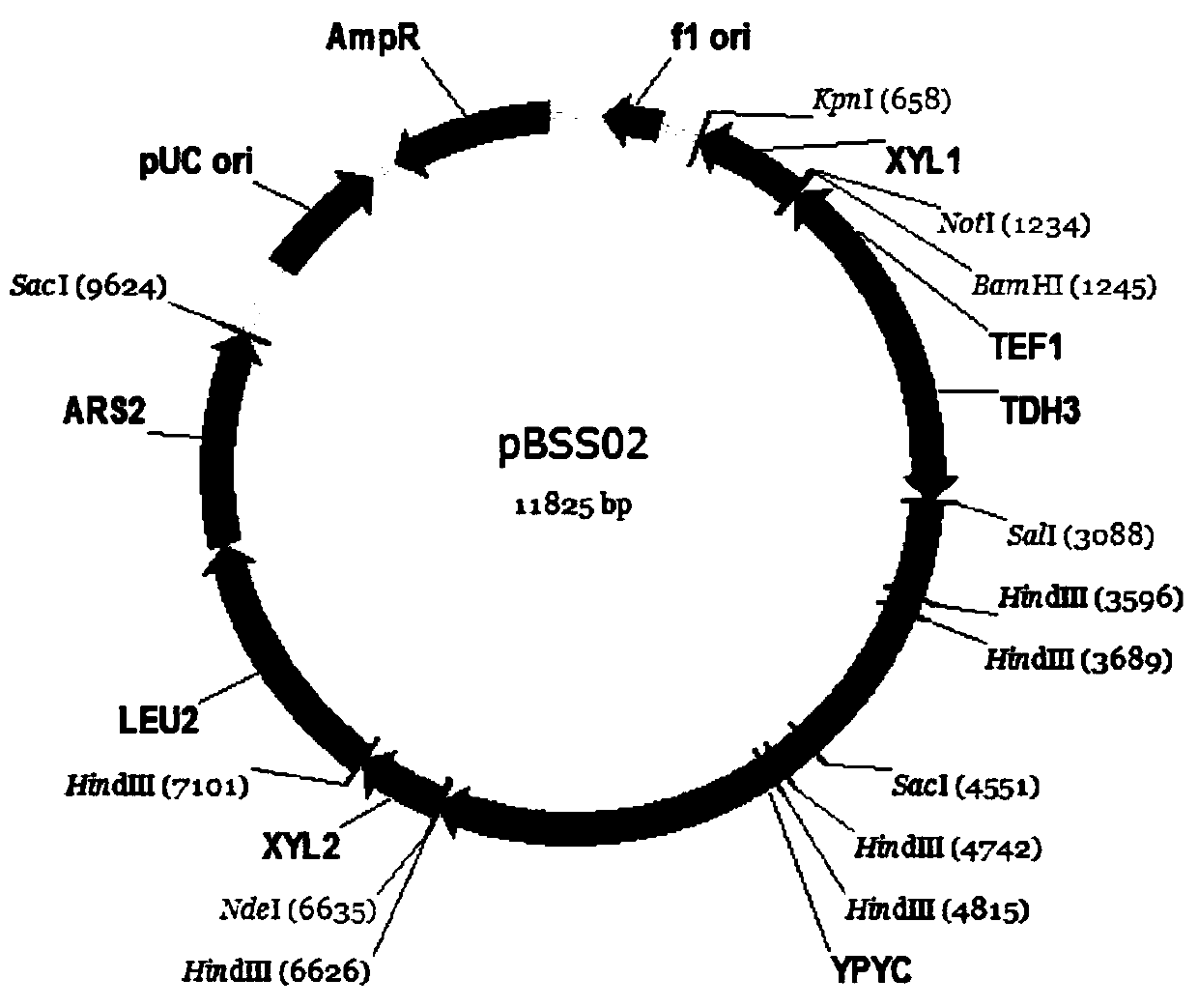
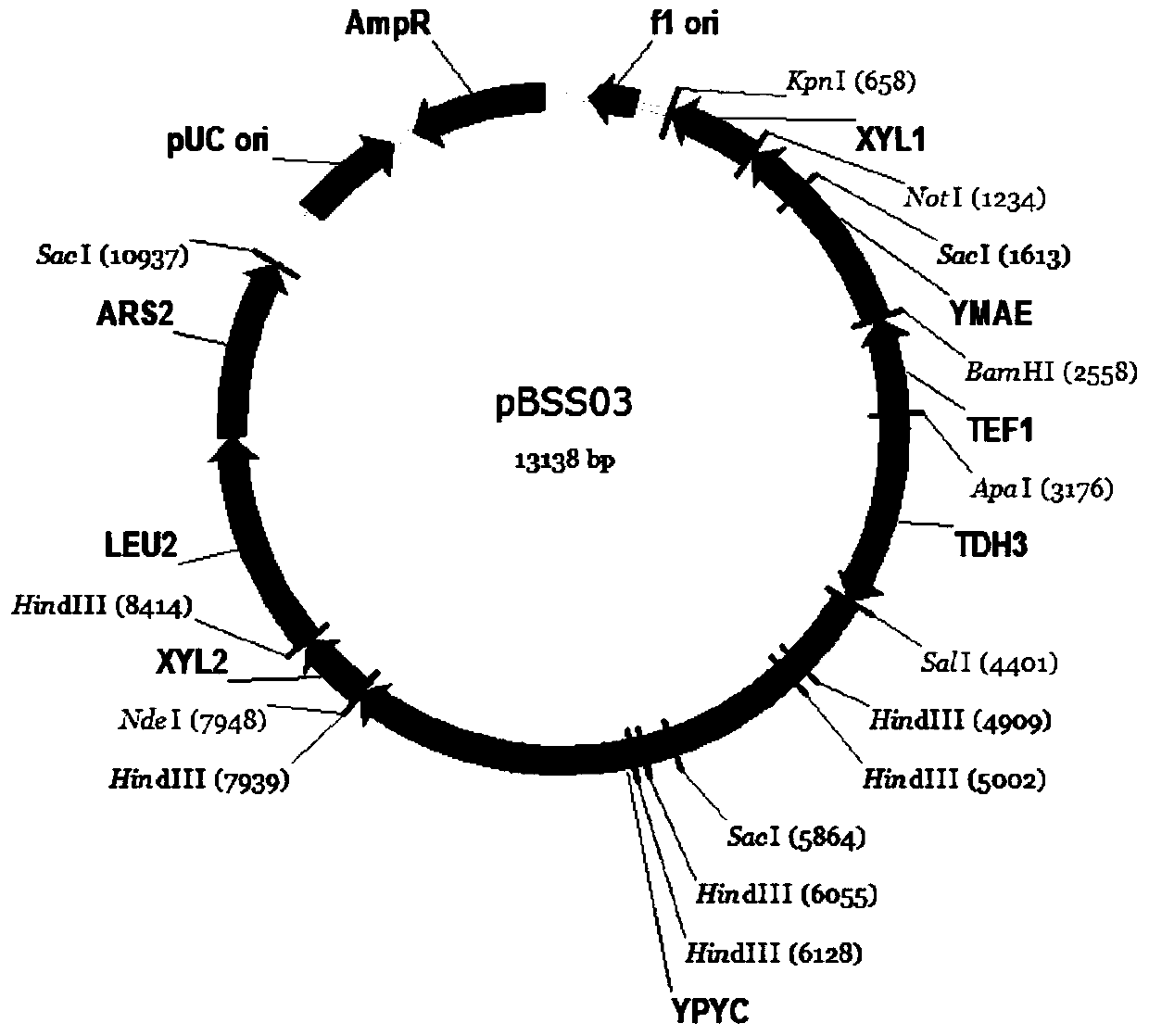
A construction method based on the production of fumaric acid by fermenting xylose with a synthetic strain of Pichia stipitis
A technology of Pichia stipitis and fumaric acid, applied in the fields of synthetic biology and bioenergy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Construction of uracil and leucine auxotrophs
[0033] (1) Using the S. stipitis genome as a template, using primers URA3-5F / URA3-5R, URA3-3F / URA3-3R, the sequences of which are SEQ ID NO: 28 and SEQ ID NO: 29, PCR amplified uracil respectively The upstream and downstream homologous fragments URAF and URAR of the key gene URA3 gene were synthesized, and a homologous recombination knockout cassette was obtained by fusion PCR. Homologous integration in yeast knocked out URA3, screened uracil-deficient strains on a screening medium containing 5-FOA, and finally obtained uracil-auxotrophic strains were verified by PCR.
[0034] (2) Using the S. stipitis genome as a template, using primers LEU2-5F / LEU2-5R, LEU2-3F / LEU2-3R, the sequences of which are SEQ ID NO: 31 and SEQ ID NO: 32, respectively PCR amplified to obtain LEU2 The upstream and downstream homologous fragments LEU2F and LEU2R of the gene are connected with URA3 with the loxP site sequence by fusion PCR ...
Embodiment 2
[0037] The knockout of embodiment 2 fumarase gene
[0038] (1) On the basis of uracil and leucine auxotrophic strains, using the S.stipitis genome as a template, design primers FUM1-5F / FUM1-5R, FUM1-3F / FUM1-3R, the sequence of which is SEQ ID NO : 35 and SEQ ID NO: 36, the upstream and downstream homologous fragments FUM1F and FUM1R of the PSfum1 gene were amplified by PCR respectively, and connected with URA3 with the loxP site sequence by fusion PCR to form a PSfum1 gene homologous recombination knockout box , PSfum1 was knocked out by homologous integration in yeast, and PSfum1-deficient strains were identified by screening on SD solid medium. The plasmid pSSH02 with Cre recombinase was transformed to remove the marker gene URA3, and a PSfum1-deficient strain was obtained. The obtained strains were verified by PCR detection.
[0039] (2) On the basis of the PSfum1-deficient strain obtained in (1), using the S. stipitis genome as a template, design primers FUM2-5F / FUM2-5R,...
Embodiment 3
[0043] The construction of embodiment 3 vector plasmids pYSS01, pBSS01 and pSSH02
[0044] (1) Extract the S.stipitis CBS6054 genome template
[0045] S.stipitis CBS6054 was cultured in YPD medium (20-25g / L peptone, 10-20g / L yeast powder, 20-25g / L glucose) at 30°C and 150-200rpm for 40-48 hours, then the genome extraction kit ( Tiangen) to extract total DNA.
[0046] (2) Using the S.stipitis CBS6054 genome as a template, design primers URA3-F / URA3-R, whose sequence is SEQ ID NO: 17, and use PCR to obtain gene URA3, whose sequence is SEQ ID NO: 11, using ApaI and NdeI double enzymes After cleavage, it was connected to the vector pY26TEF-GPD to obtain the plasmid pYSS; primers TDH3-1F / TDH3-1R and TEF1-1F / TEF1-1R were designed with the S.stipitis CBS6054 genome as a template, and their sequences were SEQ ID NO: 15 and SEQ ID NO: 16, the constitutive strong promoters TDH3p and TEF1p were respectively amplified, and the sequences thereof were SEQ ID NO: 7 and SEQ ID NO: 8. The f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com