Compositions and methods for negative selection of non-desired nucleic acid sequences
一种核酸序列、非期望的技术,应用在生物化学设备和方法、组合化学、重组DNA技术等方向,能够解决样品群体失真、不能用、丢失等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
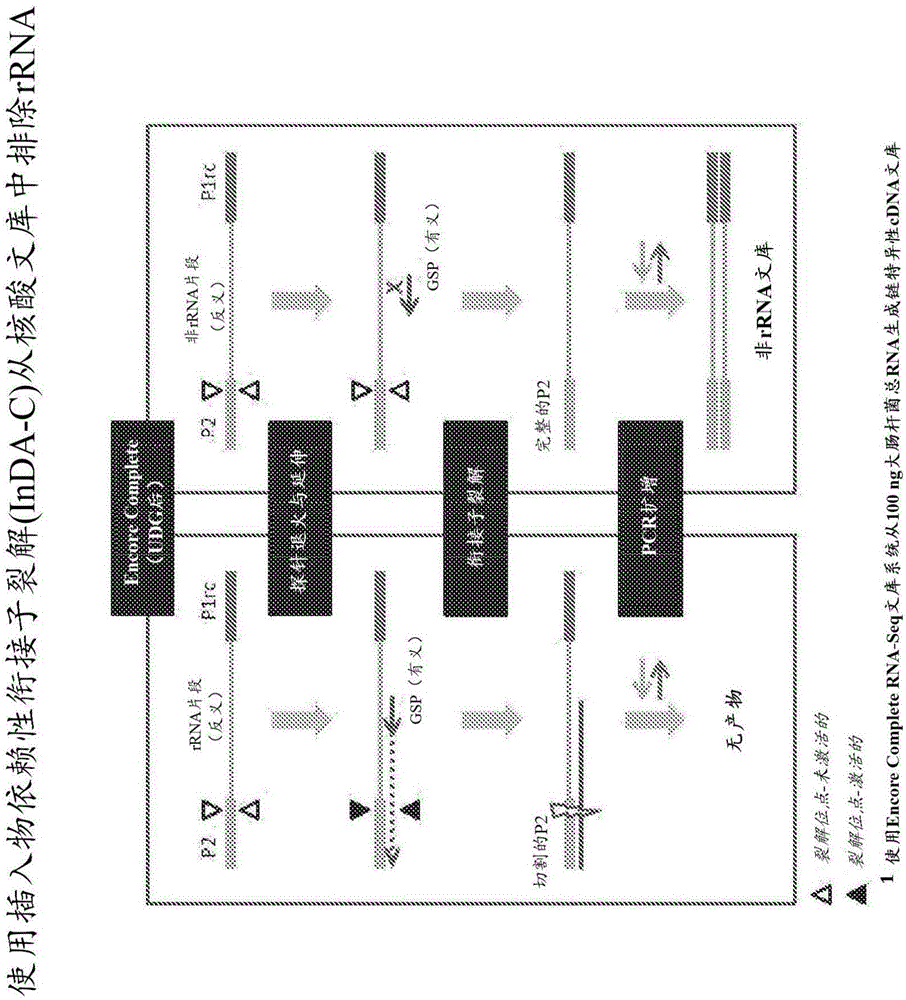
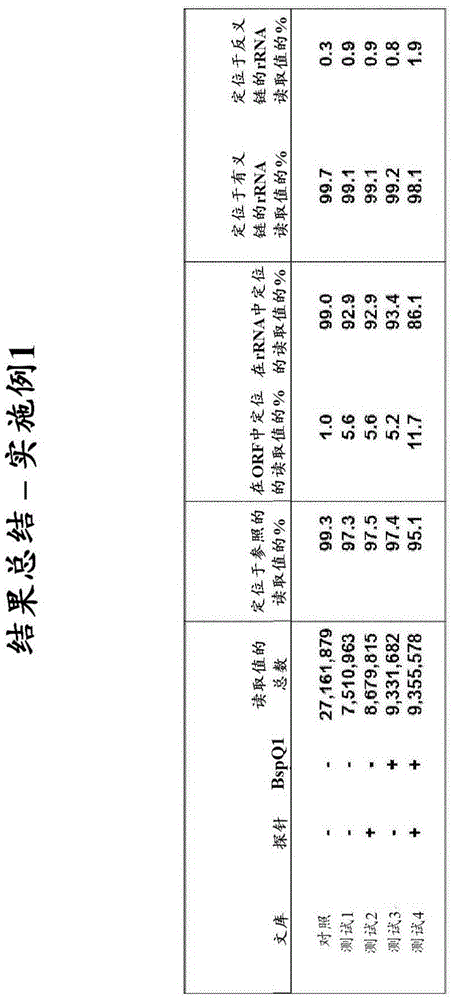
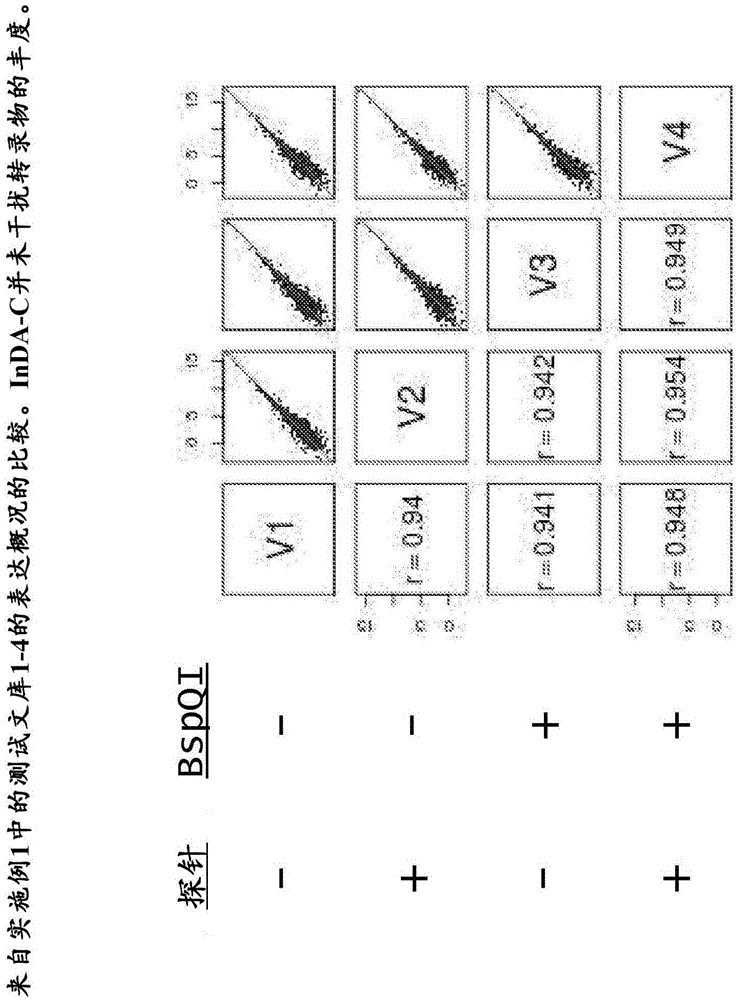
[0219] Example 1 - Exclusion of bacterial ribosomal RNA fragments from directed (ie, strand-specific) whole-transcriptome libraries.
[0220] This example describes the exclusion of four directed cDNA libraries generated from total RNA from E. Bacterial rRNA fragments.
[0221] Probe Design and Synthesis
[0222]By comparing ribosomal operons from a phylogenetically distinct set of 40 bacterial strains and 10 archaeal strains using the ClustalW multiple sequence alignment program (European Bioinformatics Institute) InDA-C probe for prokaryotic rRNA transcripts. Candidate primer sequences were first selected from highly conserved sequences identified in the 16S rRNA (9 sites) and 23S rRNA (7 sites) subunits. These conserved regions were computationally fragmented and analyzed by Primer3 (Steve Rozen and Helen J. Skaletsky (2000), Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds.) Bioinformatics Methods and Protocols: Method...
Embodiment 2
[0228] Example 2 - Exclusion of Mitochondrial DNA Fragments from Genomic DNA Libraries.
[0229] This example describes the exclusion of mitochondrial DNA fragments from genomic DNA libraries using an Insert-Dependent Adapter Cleavage (InDA-C) probe targeting the mitochondrial genome.
[0230] Probe Design and Synthesis
[0231] InDA-C was selected for annealing to both strands of the hg19 version of the human mitochondrial genomic sequence within mitochondria-specific segments identified by the "Duke 20bp uniqueness" tracks provided by the UCSC Genome Browser probe. These sequences were then screened for the best predicted melting temperature and length. Oligonucleotides ranging in length from 20-25 nt were synthesized individually and combined in equimolar ratios. The resulting probe mix was diluted to 25 times the final concentration used in the InDA-C exclusion reaction (375 nM for each species, 15 nM final).
[0232] Genomic DNA library generation
[0233] DNA li...
Embodiment 3
[0236] Example 3 - Generation of Directed cDNA Libraries ( Figure 5 ).
[0237] This example describes the generation of a directional cDNA library using conventional blunt-end ligation starting with modified duplex adapters and 50 ng of poly(A)+ selected messenger RNA.
[0238] first strand synthesis
[0239] First-strand cDNA was generated using random hexamer priming. Use containing 10μM random hexamers, 3.0mM MgCl 2 The first-strand synthesis reaction was performed with Invitrogen SuperScript III Reverse Transcriptase Kit with 1.0 mM dNTP. cDNA synthesis reactions were performed in 10 μL volumes, incubated at 40°C for 60 min and cooled to 4°C.
[0240] Second strand synthesis utilizing dUTP incorporation
[0241] Second strand synthesis was performed using the New England Biolabs NEBNext Second Strand Synthesis Module in which the second strand synthesis (dNTP-free) reaction buffer was supplemented with a dNTP mix containing 0.2 mM dATP, dCTP and dGTP and 0.54 mM d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com