A PCR identification method between the germplasm of flounder and its close relative alien species
A technology of brown flounder and close relatives, applied in the field of PCR identification, can solve the problems of unseen species identification technology of brown flounder and Atlantic flounder, and few patents for detection and identification of biological germplasm resources, and achieve simple technology Ease of operation, speed up the detection process, and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] 1. Mitochondrial genomic DNA extraction:
[0024] Using the back muscle material of brown flounder and its close relative species Atlantic flounder as raw materials, take 100 mg each and treat them with sterile water, grind them into powder in liquid nitrogen, put them into a 1.5ml centrifuge tube, Refer to the refined version of the Molecular Cloning Experiment Guide (J. Sambrook, D.W. Russell) and use the standard phenol / chloroform extraction method to extract the mitochondrial genomic DNA from the raw materials, and set them aside.
[0025] 2. Specific primer design:
[0026] Based on the complete mitochondrial sequences and partial control region sequences of flounder and Atlantic flounder published in the GENBANK database, DNASTAR8.0 software was used for homologous sequence comparison, and PRIMER5.0 software was used to assist in the design of specific primers.
[0027] 2.1 Amplification primers:
[0028] (1) The name and sequence of the amplification primers fo...
Embodiment 2
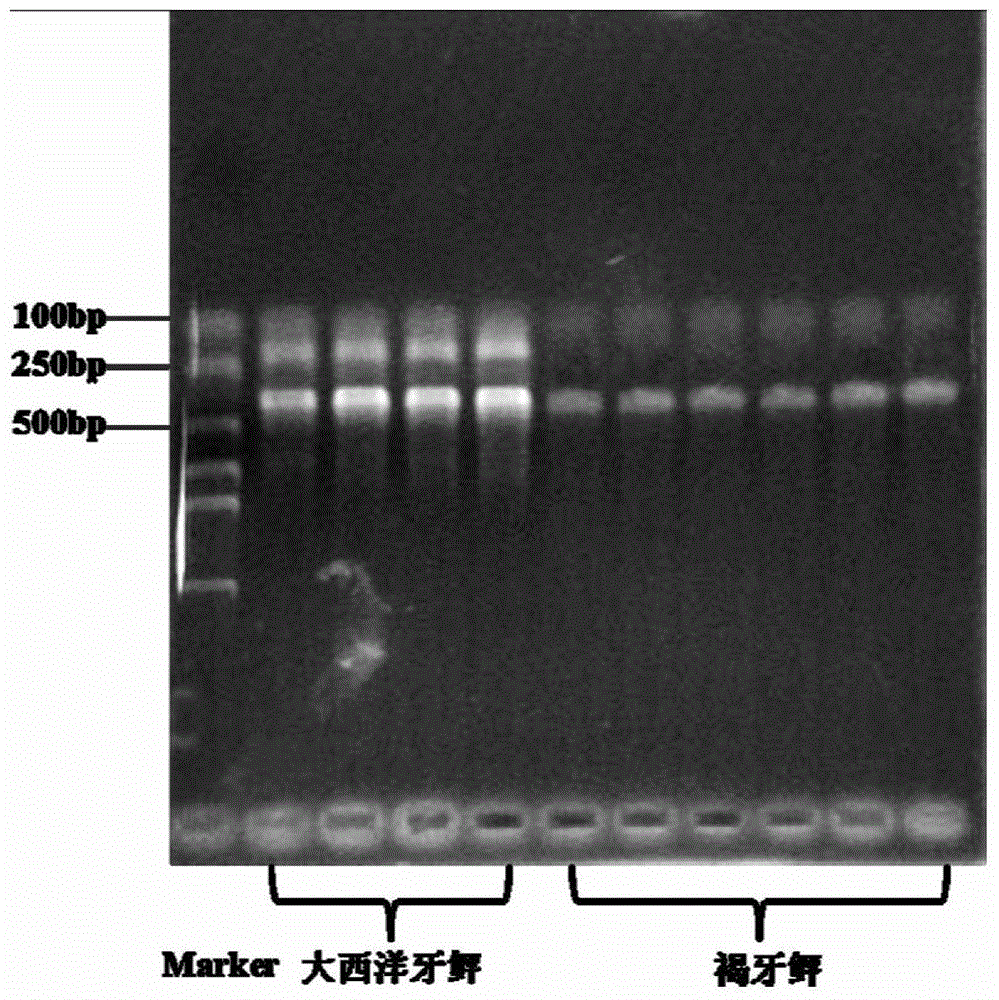
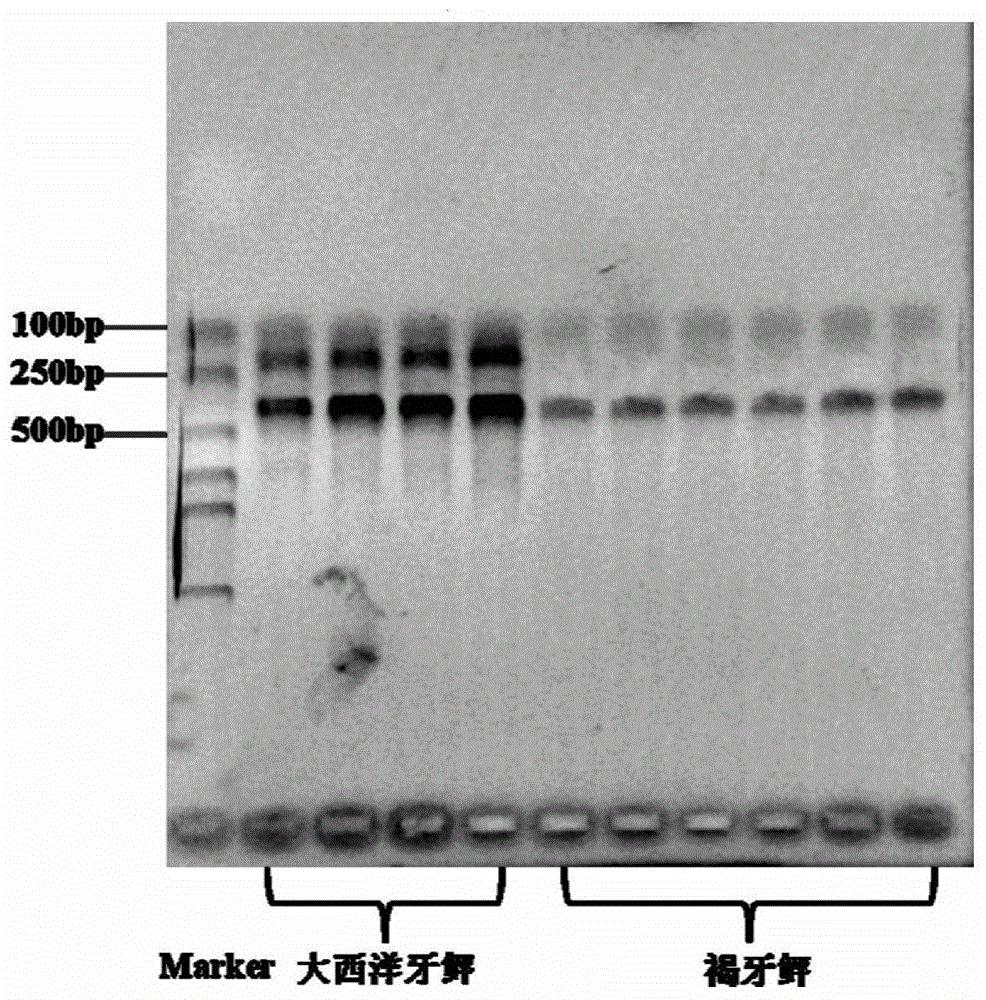
[0048] Using the DNA of the back muscle of the juvenile flounder and its close relative alien species Atlantic flounder as a template, based on the PCR reaction conditions in Example 1, and using the common cultured flounder species turbot as a negative control, the PCR reaction The results showed that the Atlantic flounder sample had specific amplification positive bands at 422bp and 201bp, the brown flounder sample had a positive amplification band at 422bp, and the turbot sample had no corresponding specific band (see figure 1 ).
[0049] From the above, it can be known that the F / R of the primer of the mitochondrial DNA control region 1 The combination can amplify both Atlantic flounder and brown flounder, primers F / R 2 The combination only had a species-specific amplification effect on Atlantic flounder, so there were differences in PCR amplification bands between brown flounder and Atlantic flounder, while F / R 1 and F / R 2 The combination is non-specific for other flo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com