Method and device for carrying out GC correction on chromosome sequencing results
A chromosome and sequence information technology, applied in the field of fetal genetic abnormality detection, can solve the problems of interference detection accuracy, difficulty, data distortion, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0119] Example 1 Analysis of Factors Affecting Detection Sensitivity: GC Bias and Gender
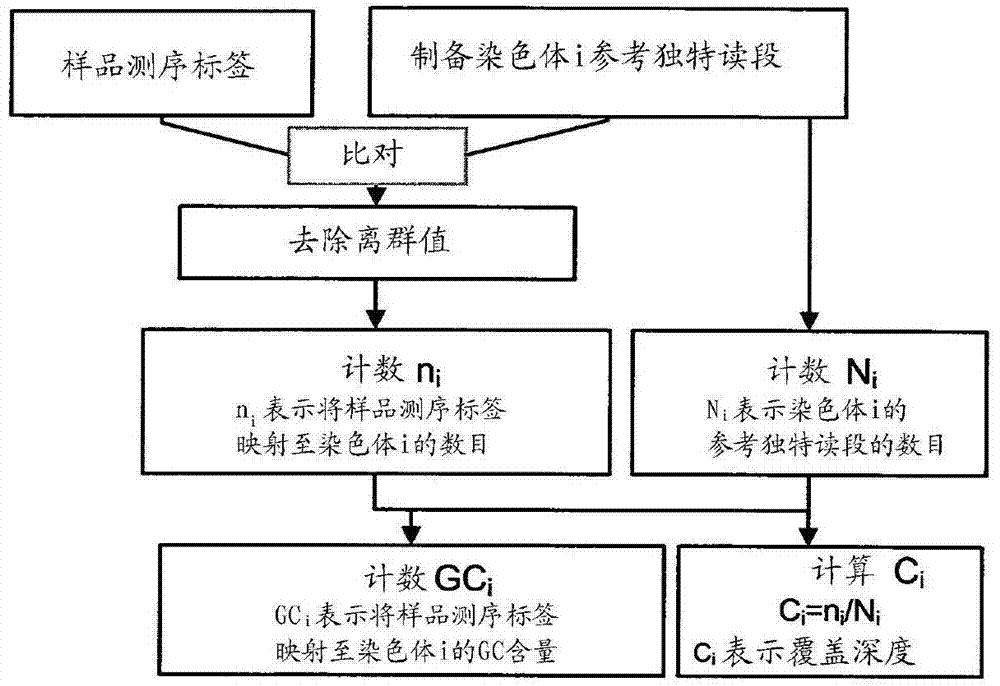
[0120] A schematic framework of steps for calculating coverage depth and GC content is shown in figure 1 . The inventors used software to generate reference unique reads by cutting the hg18 reference sequence into l-mers (here l-mers are the reads that were artificially dissected from the human sequence reference with the same length "l" as the sample sequencing reads) , collect these "unique" l-mers as reference unique reads for inventors. Second, the inventors mapped their sequencing sample reads to reference unique reads for each chromosome. Third, the inventors removed outliers by applying a quintile outlier cutoff to obtain a clean dataset. Finally, the inventors calculated the coverage depth for each chromosome for each sample, and calculated for each sample the GC content of the sequenced unique reads that mapped to each chromosome.
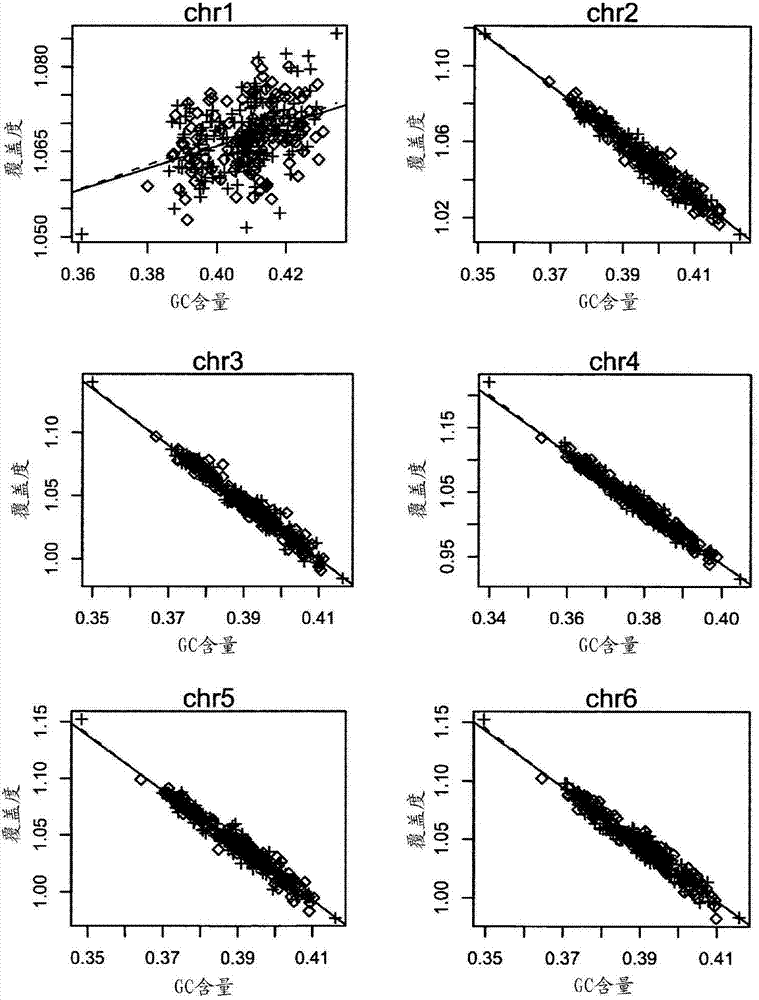
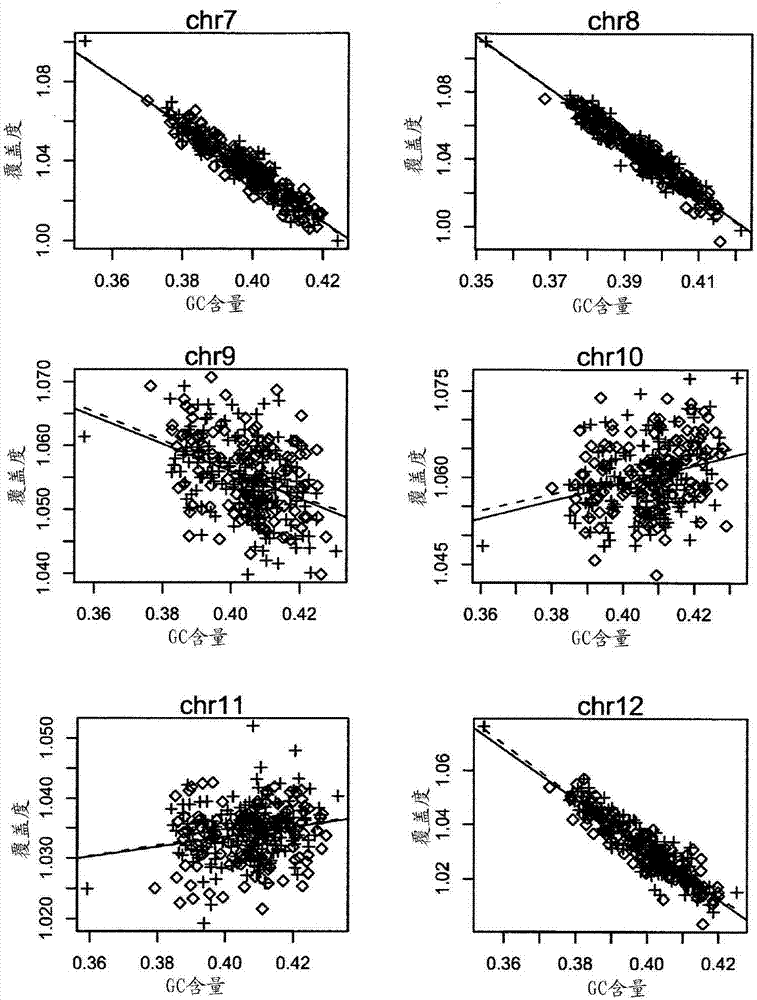
[0121] To investigate how GC content af...
Embodiment 2
[0124] Embodiment 2 statistical model
[0125] Using this phenomenon discussed above, the inventors attempted to fit the relationship between coverage depth and corresponding GC content using local polynomials. Coverage depth is composed of the following function of GC and normally distributed residuals:
[0126] cr i,j =f(GC i,j )+ε i,j ,j=1,2,...,22,X,Y (4)
[0127] where f(GC i,j ) represents the relationship between the coverage depth of sample i, chromosome j and the corresponding GC content, ε i,j Represents the residual of sample i and chromosome j. There is not a strong linear relationship between the depth of coverage and the corresponding GC content, so the inventors applied the loess algorithm to fit the depth of coverage to the corresponding GC content, from which the inventors calculated the Say the important value, namely the fit coverage depth:
[0128] cr i , j ...
Embodiment 3
[0132] Example 3 Fetal Fraction Estimation
[0133] Since fetal fraction is so important to the inventor's assay, the inventor estimated the fetal fraction prior to the testing procedure. As noted above, the inventors sequenced 19 adult males, and when comparing their coverage depths with those of female fetuses, the inventors found that the chromosome X coverage depth in males was nearly 1 / 2 that of females, and that chromosome X in males The depth of Y coverage is approximately 0.5 times greater than that of females. Therefore, the inventors can rely on the coverage depth of chromosomes X and Y and consider GC correlations to estimate the fetal fraction as Equation 8, Equation 9 and Equation 10:
[0134] fy i = ( cr i , Y - c ^ ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com