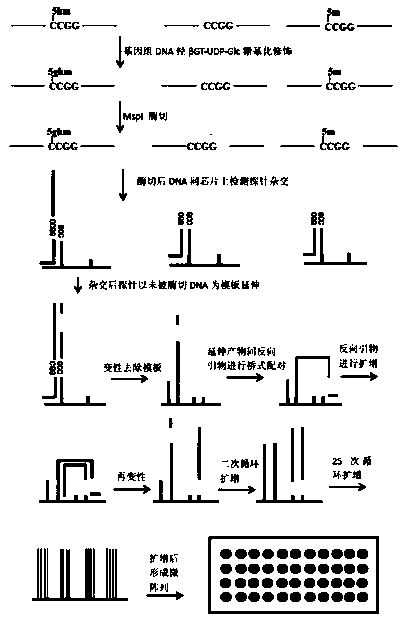
Bridge-PCR-based method for detecting DNA hydroxymethylation
A hydroxymethylation and bridging technology, applied in the field of high-throughput and high-sensitivity detection, can solve the problem of difficult to accurately analyze the detection cost of specific base 5hmC, and achieve the effects of high repeatability, reduced test cost and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Realized the detection of 10 CCGG sites (32131-40796) hmC in the spinal cord BDNF gene (Genbank ACCESSION: NT_039207) of mice with chronic inflammatory pain.
[0026] For the above 10 CCGG sites, design corresponding forward and reverse amplification primers for each site (the reverse primer is 27 bases long: the 7 bases near the 5' end are T, and the 20 bases near the 3' end are T The sequence after CGG (including CGG) of each CCGG site is reverse complementary. The amplification length of forward and reverse primers is 120bp;
[0027] The nucleotide sequences of the forward and reverse primer pairs of the above 10 CCGG sites are as follows:
[0028] (1) 32048-32167 F1 (T) 7 TACTGGGGCATATAAAGTT R1 (T) 7 ATGAACTAACCAGTACCCCG
[0029] (2) 32198-32317 F2 (T) 7 CCTTTAGCTCCTTGGCTACT R2 (T) 7 TCTCTTGTGAGACTATGCCG
[0030] (3) 32808-32927 F3 (T) 7 TATGAACATAGTGGAGCATG R3 (T) 7 AATTGGACATAGTACTACCG
[0031] (4) 34100-34219 F4 (T) 7 TACTGGGGCATATAAAGTT R4 (T) 7 GCTGA...
Embodiment 2
[0041] Achieved 9 CCGG sites (1222-3705) hmC of BDNF gene (Genbank ACCESSION: NC_000011.9) and 8 CCGG sites (771-15422) hmC of COMT gene (Genbank ACCESSION: NT_187012.1) in plasma DNA of patients with chronic rheumatoid pain simultaneous detection.
[0042] For the above 12 CCGG sites, design corresponding forward and reverse amplification primers for each site: the reverse primer is 27 bases long: the 7 bases near the 5' end are T, and the 20 bases near the 3' end are T The sequence after CGG (including CGG) of each CCGG site is reverse complementary. The amplification length of the forward and reverse primers is 120bp; the length of the forward primers is 20bp, and the 5' ends of the forward and reverse primers are both acryloyl-modified. Each pair of primers is diluted and mixed to 20uM, and fixed on the slide treated with acrylamide called a microarray chip.
[0043] The forward and reverse primer sequences for BDNF gene detection are as follows:
[0044] (1) 1222-1341 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com