RNA interference preparation used for treating viral hepatitis B
A hairpin-type technology for hepatitis B virus, applied in antiviral agents, DNA/RNA fragments, viruses, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
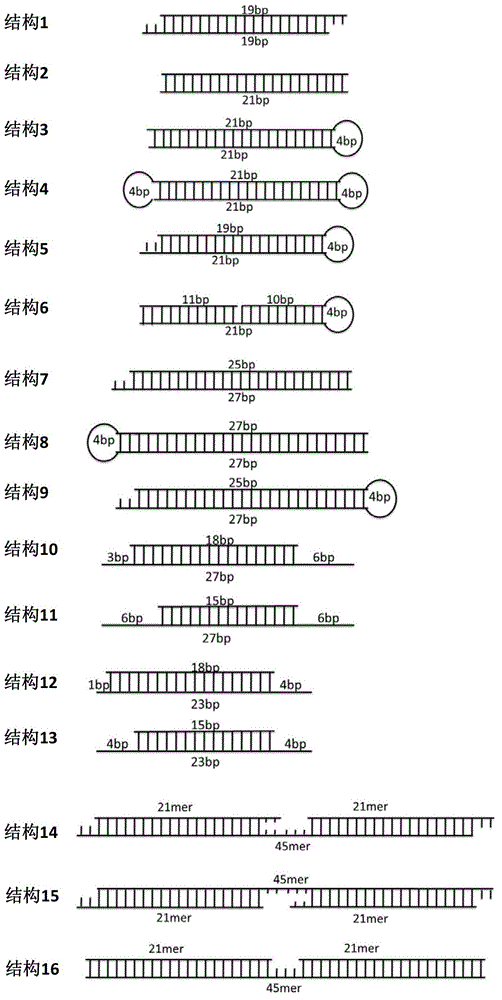
[0083] The segmented iNA includes antisense strands targeting two different HBV RNA loci. There are several non-complementary nucleotides in the middle of the continuous strand, for example:
[0084] iNA sequence number 231 (iNA ID#2and#8):
[0085] Sense strand: GGGUUUUUCUUGUUGACAAdTdTuauaCCGUGUGCACUUCGCUUCAdTdT
[0086] Antisense strand 1: UUGUCAACAAGAAAAACCCCG
[0087] Antisense strand 2: UGAAGCGAAGUGCACACGGUC
[0088] iNA ID#231A (segmented iNA designed according to iNA ID#2and#8iNA):
[0089] Sense strand: CCGUGUGCACUUCGCUUCAdTdTuauaGGGUUUUUCUUGUUGACAAdTdT
[0090] Ant i-sense strand1: UUGUCAACAAGAAAAACCCCG
[0091] Anti-sense strand2: U GAAGCGAAGU GCACACGGUC
[0092] iNA ID#232 (Segmented iNA designed according to iNA ID#2 and #5iNA)
[0093] Sense strand: GGGUUUUUCUUGUUGACAAdTdTuauaGGAUGUGUCUGCGGCGUUUUdTdT
[0094] Ant i-sense strand1: UUGUCAACAAGAAAAACCCCG
[0095] Ant i-sense strand2: AAACGCCGCAGACACAUCCAG
[0096] iNA ID#233 (segmented design according to i...
Embodiment 2
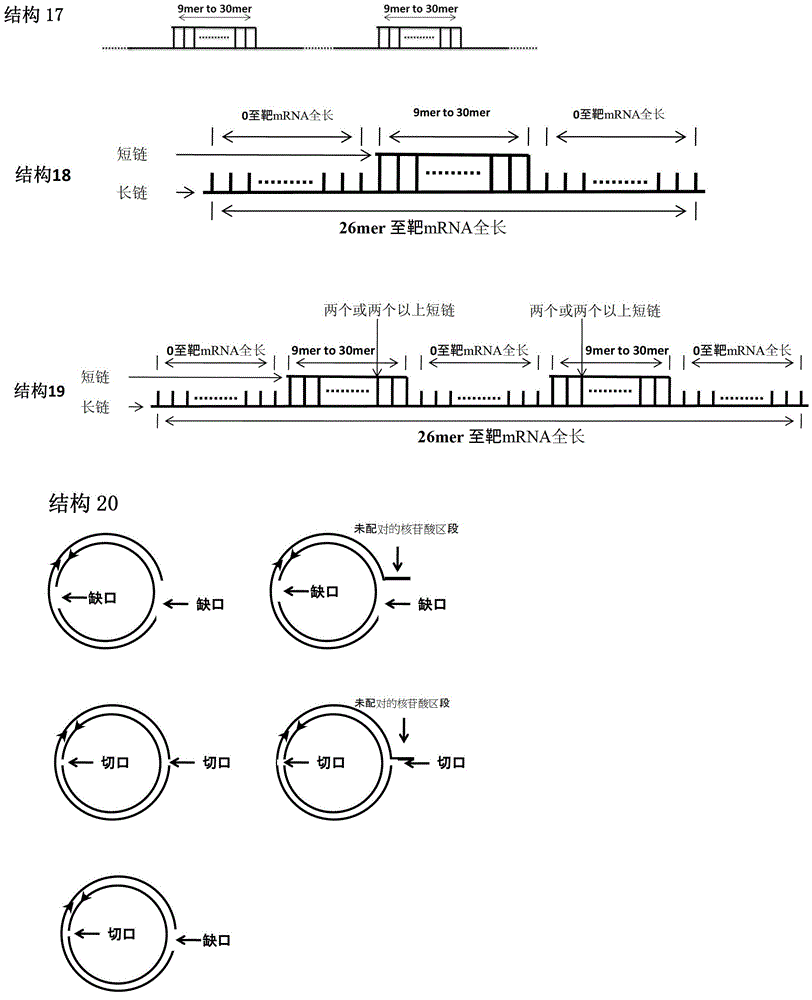
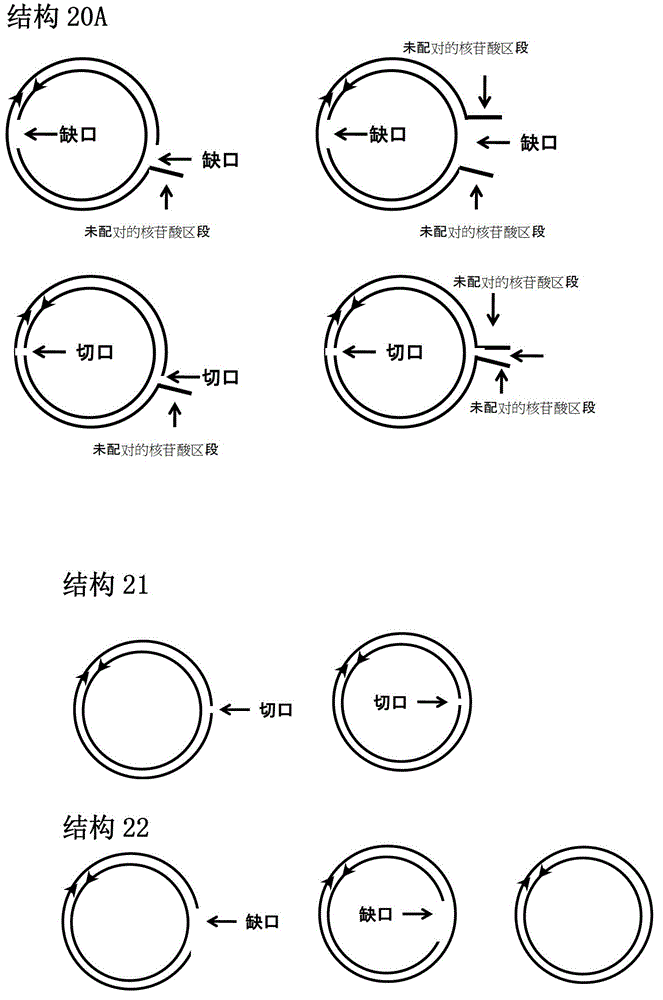
[0133] For ring or ring-like structures, each iNA is as follows. One strand consists of two polynucleotides in opposite directions (one 5' to 3' and the other 3' to 5'), where the 5'-3' direction is in the same order as the sequence of the target RNA. In solution, the polynucleotides at the two ends are hybridized together to form a circular structure. The other strand will be complementary to the single-stranded region of the loop and also to the target RNA. An example of such a sequence is as follows:
[0134] iNA ID#242 (circle-like HBV iNA designed according to iNA ID#2):
[0135] Justice Chain: GGGCCCGGGUUUUUCUUGUUGACAAUU CCCGGG (The underlined part is 3'-5' direction)
[0136] Antisense strand: UUGUCAACAAGAAAAACCCCG
[0137] iNA ID#243 (circle-like HBV iNA designed according to iNA ID#5):
[0138] Justice Chain: GGGCCCGGAUGUGUCUGCGGCGUUUUU CCCGGG (The underlined part is 3'-5' direction)
[0139] Antisense strand: AAACGCCGCAGACACAUCCAG
[0140] iNA ID#244 (quas...
Embodiment approach 3
[0156] Chemical modification:
[0157] iNA ID#249 (circle-like HBV iNA designed by iNA ID#2 and #8)
[0158] Sense strand: mGGGUUUUUCUUGUUGAmCAAdTdTuauamCCGmUGUGmCACUUCGCUUmCAdTdT
[0159] Antisense strand 1: UUGUCAACAAGAAAAACCCCG
[0160] Antisense strand 2: UGAAGCGAAGUGCACACGGUC
[0161] (bases before "m" refer to 2'-methoxy (2'-OMethyl) RNA bases. Bases before "d" refer to DNA bases)
[0162] iNA ID#250 (segmented design according to iNA ID#2 and #8iNA):
[0163] Sense strand: mGGGUUUUUCUUGUUGAmCAAdTdTuauamCCGmUGUGmCACUUCGCUUmCAdTdT
[0164] Antisense strand 1: UUGUmCAAmCAAGAAAAACCCCG
[0165] Antisense strand 2: UGAAGCGAAGUGCACACGGUC
[0166] (bases before "m" refer to 2'-methoxy (2'-O-Methyl) RNA bases. Bases before "d" refer to DNA bases)
[0167] iNA ID#250A (segmented design according to iNA ID#2 and #8iNA):
[0168] Sense strand: mCCGmUGUGmCACUUCGCUUmCAdTdTuauamGGGUUUUUCUUGUUGAmCAAdTdT
[0169] Antisense strand 1: UUGUmCAAmCAAGAAAAACCCCG
[0170] Antisense s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com