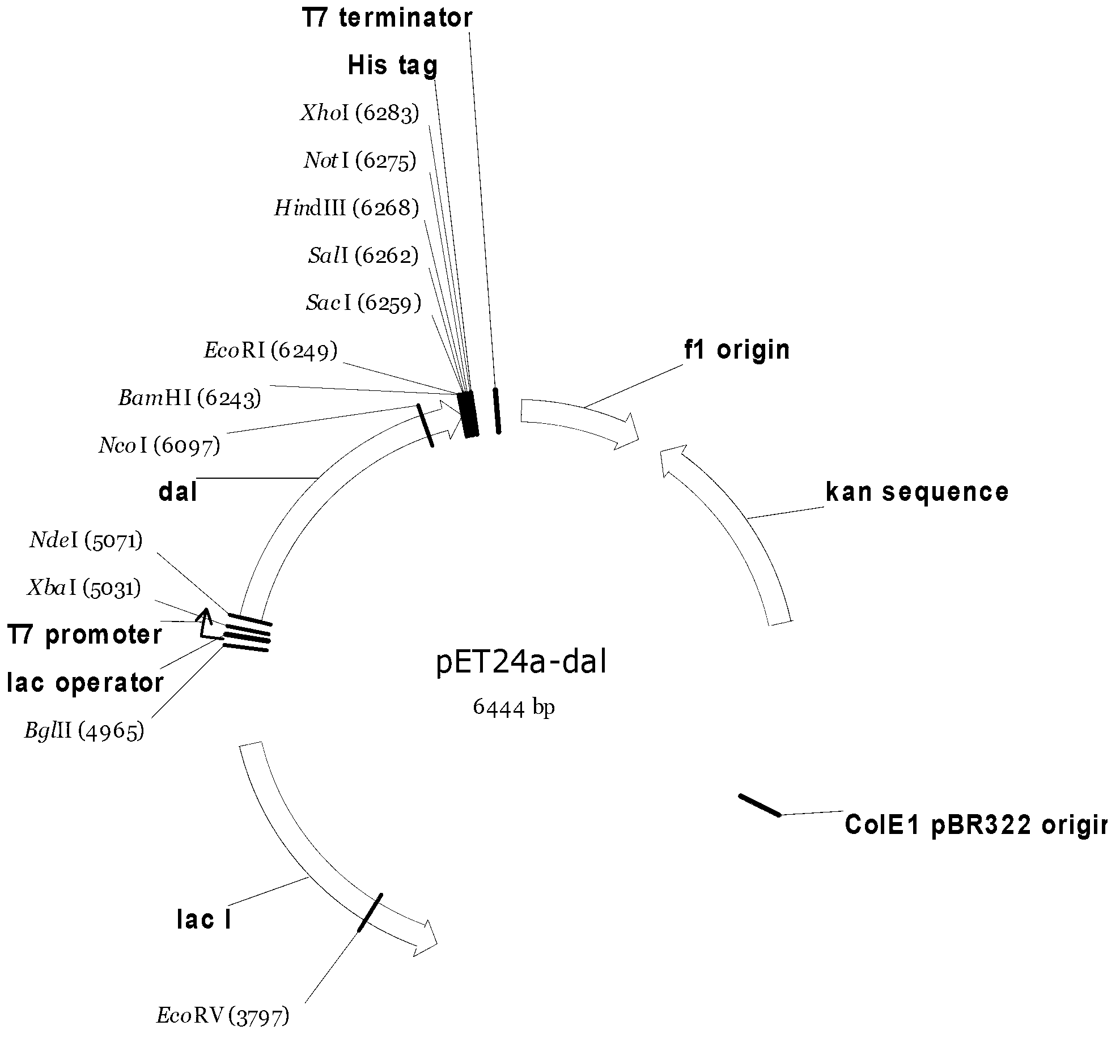
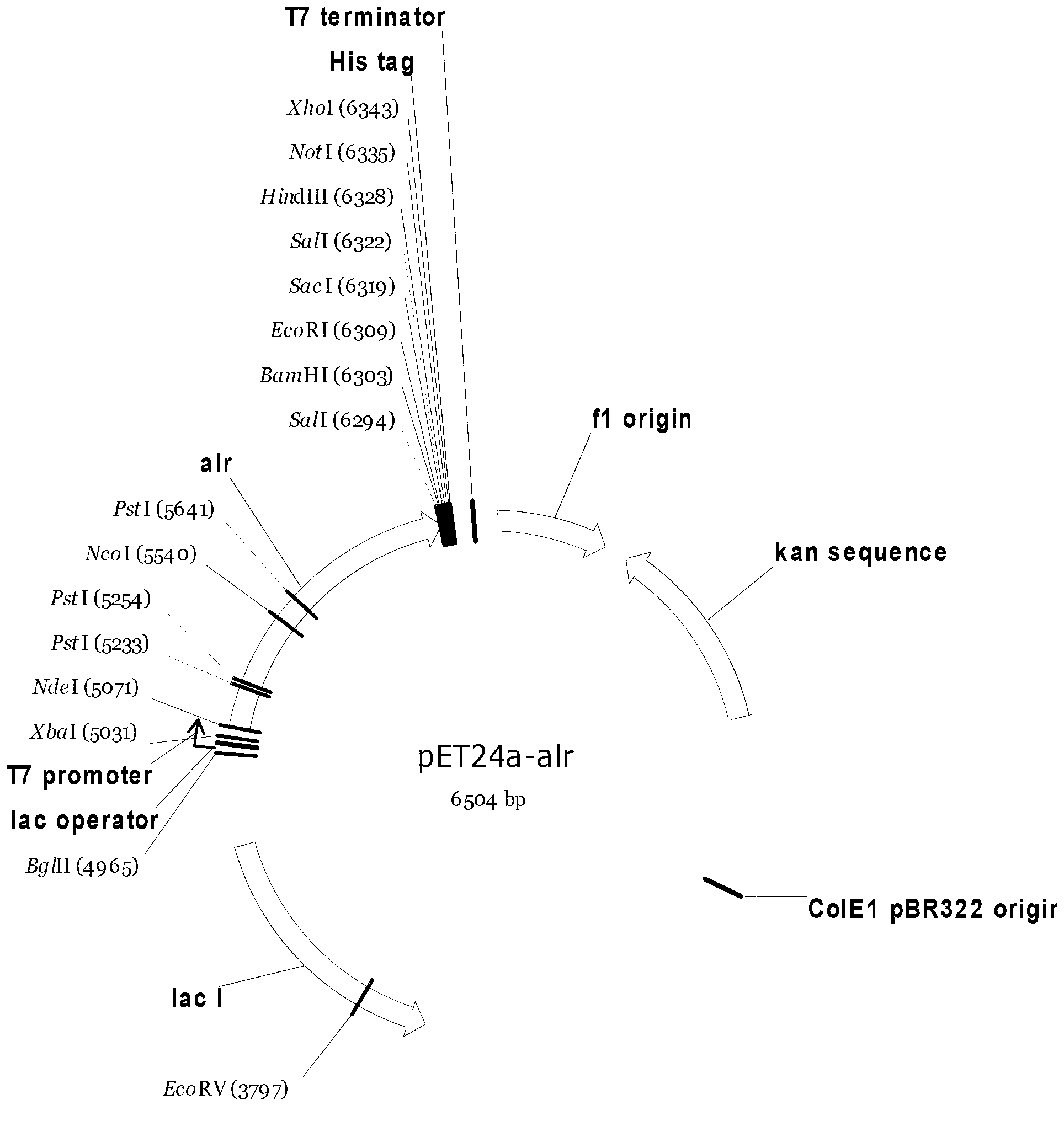
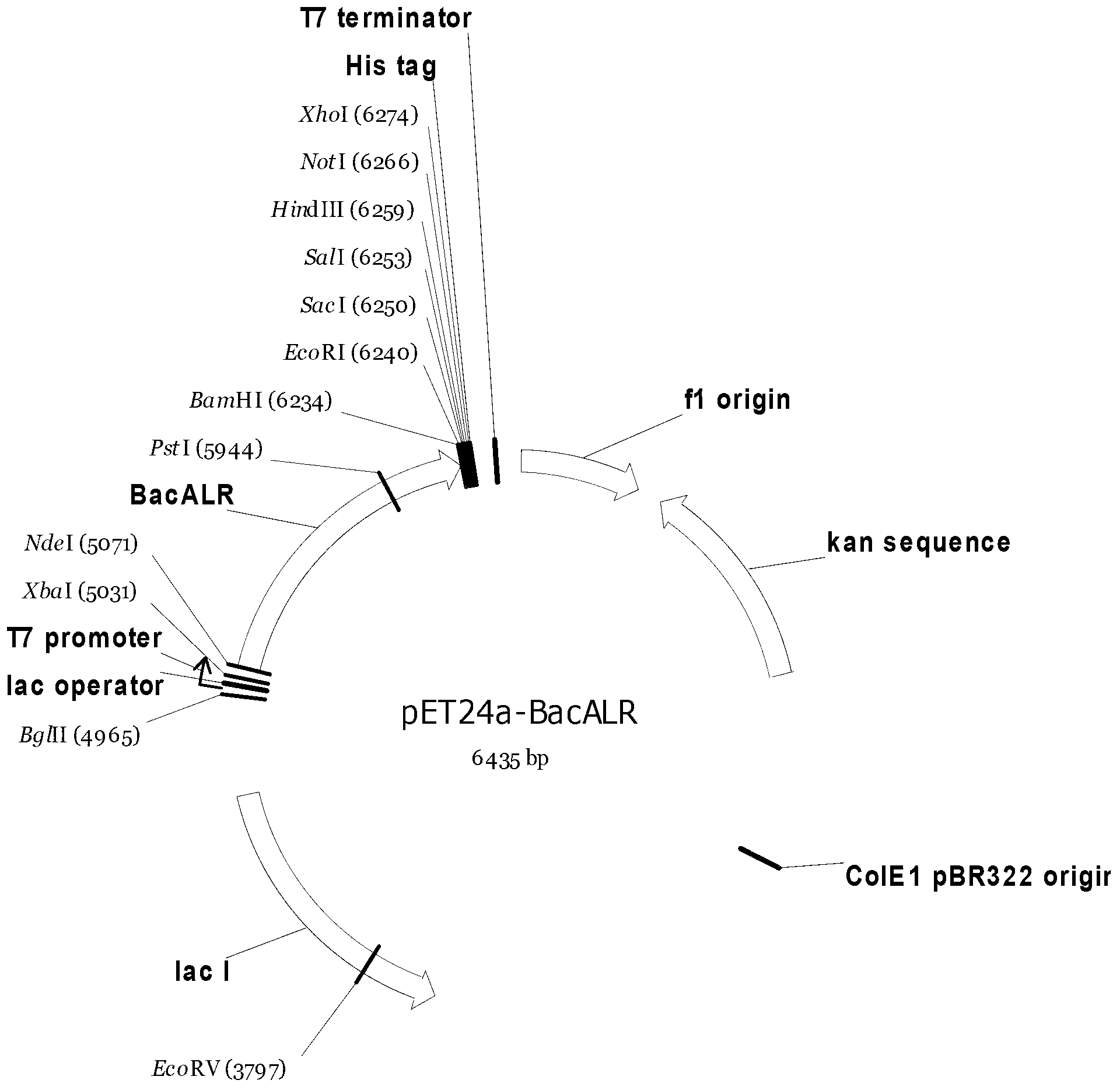
Strain for producing high-yield DL-alanine and application thereof
A technology of alanine and alanine racemase, applied in bacteria, microorganisms, recombinant DNA technology, etc., can solve the problem of lack of efficient production of alanine racemase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0086] The expression strain provided by the invention can be efficiently applied to the production of DL-alanine. In a preferred example of the present invention, the preparation method of said DL-alanine comprises steps:
[0087] Transform raw material L-alanine with the bacterial strain provided by the invention to obtain DL-alanine;
[0088] Alternatively, the method comprises: converting the raw material L-alanine with the fermentation broth of the bacterial strain provided by the present invention to obtain DL-alanine. Wherein, the fermented liquid is a fermented liquid containing bacteria.
[0089] In another preferred example, the method includes: cultivating the bacterial strain described in the present invention in the presence of L-alanine.
[0090] In another preferred example, the conversion rate of the L-alanine is ≥95%, preferably ≥98%, more preferably ≥99%.
[0091] In another preferred example, the dosage of the strain is 0.5-5 g / l, preferably 1-3 g / l.
[...
Embodiment 1
[0118] Example 1 Construction of Bacillus subtilis-derived D-alanine racemase expression strain
[0119] Bacillus subtilis subsp.subtilis str.168 was inoculated in LB liquid medium, and cultured at 220 rpm at 30°C for 24 hours. For the extraction of total DNA, refer to the instructions of the genome extraction kit.
[0120] According to the reported gene sequence of D-alanine racemase gene dal derived from Bacillus subtilis subsp.subtilis str.168 (NCBI accession number: AL009126.3), a sense primer and an antisense primer were synthesized. The nucleotide sequences of the primers are respectively recorded in SEQ ID NO: 1 (dal-NdeI-F) and SEQ ID NO: 2 (dal-BamHI-R).
[0121] SEQ ID NO: 1dal-NdeI-F
[0122] GGAATTCCATATGAGCACAAAACCTTTTTAC
[0123] SEQ ID NO: 2dal-BamHI-R
[0124] CGGGATCCTTAATTGCTTATATTTACCT
[0125] The reaction solution was subjected to PCR amplification, and 50 μL of the reaction solution contained the above-mentioned pair of primers, wherein each primer was...
Embodiment 2
[0128] Example 2 Construction of alanine racemase expression strain derived from Pseudomonas putida KT2440
[0129] The alanine racemase gene alr (NCBI accession number: NC_002947) derived from Pseudomonas putida KT2440 was synthesized to obtain the recombinant plasmid pUC57-alr.
[0130] Digest pUC57-alr with NdeI and BamHI at 37°C for 3-6 hours. The enzyme digestion system is: pUC57-alr38μL, 10X Buffer Tango10μL, NdeI1μL, BamHI1μL, nucleic acid electrophoresis and gel recovery kit to recover the 1.2kb alr fragment.
[0131] The recovered alr fragment was ligated with the expression vector pET24a treated by the same enzyme digestion with T4 DNA ligase overnight at 16°C, transformed into Escherichia coli DH5α competent cells with calcium chloride method, spread on LB plates containing Kan, and cultured overnight at 37°C . Pick a single clone and inoculate it in an LB test tube for culture, extract the plasmid with a plasmid extraction kit, and verify it by double enzyme diges...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com