Plant salt tolerance related miRNA (microribonucleic acid) and application thereof
A salt-tolerant, plant-based technology for miRNA, miRNA precursors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Embodiment 1: Salt mustard seed collection and planting
[0034] Wild-type Thellungiella salsuginea (Shandong eco-type) seeds were collected in the saline-alkali land of the Yellow River Delta in Dongying City, Shandong Province, dried in the sun, and subjected to surface disinfection: 70% alcohol, treated for 1 min, 1% NaClO solution, vortexed for 10-15 min, and sterilized After rinsing with double distilled water, they were sown in petri dishes with MS medium. The temperature setting of the light incubator was: 25°C for 16h during the day and 22°C for 8h at night. After growing 2 true leaves, transfer them to Hogland liquid medium. After culturing for about 5 weeks, add NaCl to the Hogland nutrient solution to make the final concentration 200mM, and cultivate for 24 hours. The control is still Hogland nutrient solution without NaCl.
Embodiment 2
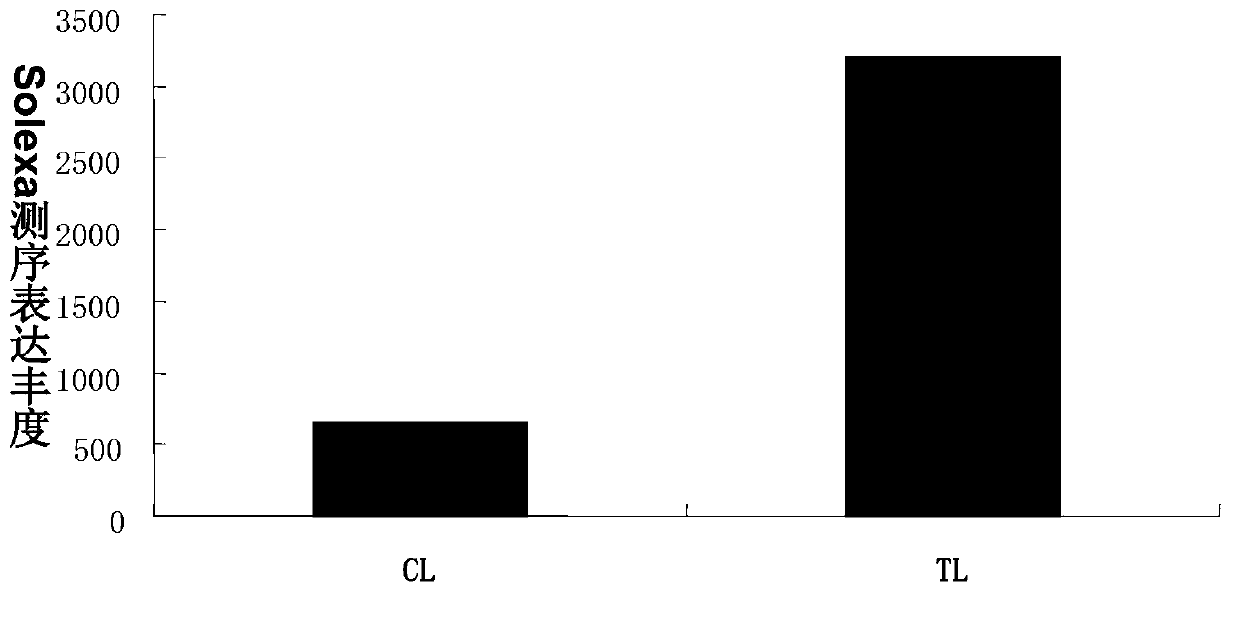
[0035] Example 2: Construction and Solexa sequencing of Salt mustard miRNA library
[0036] The samples of the salt mustard control group (CL) and the NaCl treatment group (TL) were sent to Shenzhen BGI Institute to construct the miRNAs library for Solexa sequencing. After removing 3' adapters, contaminating sequences, less than 18nt sequences and polyA sequences after sequencing, a total of 12,010,658 high-quality small RNA sequences (sRNA) were obtained in the Salina CL miRNA library, and a total of high-quality small RNA sequences were obtained in the TL miRNA library (sRNA) 12330771 entries. The result of bioinformatics prediction showed that the mature body sequence of tsa-miRn646 was UGAAGGAUCGAGGUCGAGGCA. The precursor sequence of tsa-miRn646 is AATAACAAGGTGAAGGATCGAGGTCGAGGCAGTATTGATATAGATTCAGCTCGTCCTTCACCGGTGCAAC, and its position in the genome is: scaffold_6:6637138:6637207:-. According to the precursor and mature body sequences of tsa-miRn646, use the software RNA...
Embodiment 3
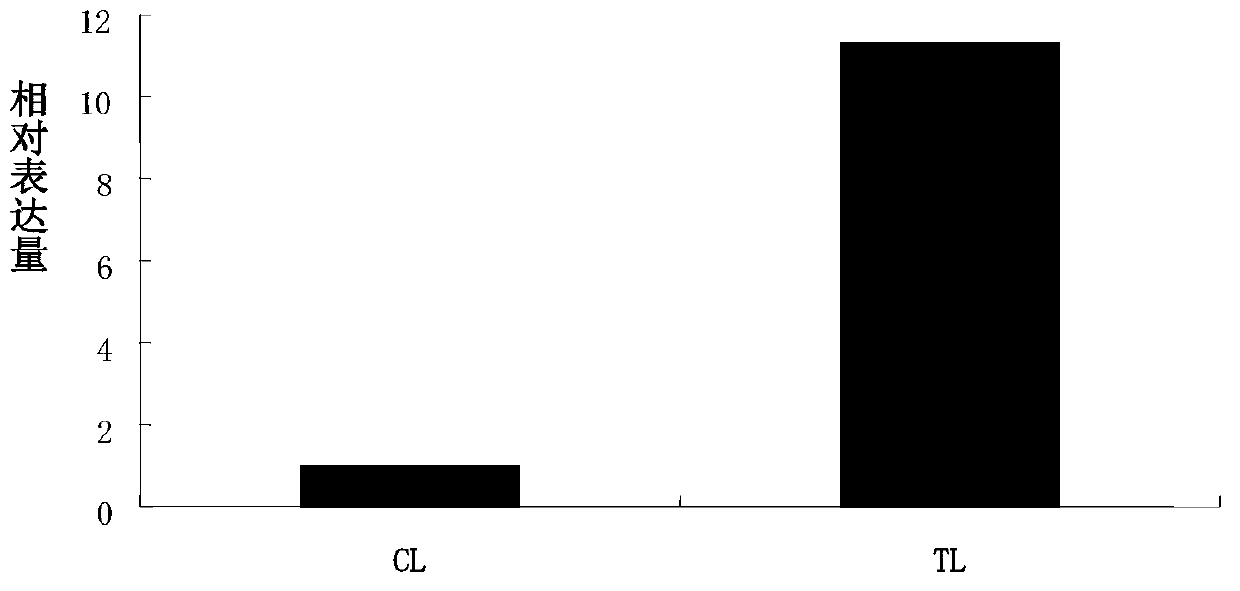
[0038] Example 3: Real-time quantitative PCR verifies the expression level of tsa-miRn646 under control and 200mM NaCl treatment
[0039] (1) Primer design for tsa-miRn646 reverse transcription and Stem-loop quantitative PCR:
[0040] According to the sequence of the mature miRNA of tsa-miRn646 and the sequence of the internal standard gene - Salt mustard U6, the primers for stem-loop PCR were designed as follows:
[0041] 1) tsa-miRn646 Stem-loop RT primer:
[0042] GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTGCCT,
[0043] 2) tsa-miRn646 Forward primer: CGGCTGAAGGATCGAGGTCG,
[0044] 3) tsa-miR n646 Reverse primer: GTGCAGGGTCCGAGGT,
[0045] 4) U6-Forward primer: GATAAAATTGGAACGATACAG,
[0046] 5) U6-Reverse primer: ATTTGGACCATTTCTCGATTT.
[0047] As shown in SEQ ID NO: 4-8 in the sequence listing.
[0048] (2) Extraction of salt mustard miRNA
[0049] The processing and collection of salt mustard samples are the same as in Example 1.
[0050] Salt mustard miRNA was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com