SNP (single nucleotide polymorphism) data filtering method
A screening method and data technology, applied in the intersection of computer and biomedicine, can solve the problems of high cost, lack of persuasiveness of SNP sites, no consideration of interaction, etc., and achieve the effect of enhancing reliability and accurate classification weight value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The present invention will be described in further detail below in conjunction with the accompanying drawings and preferred embodiments.
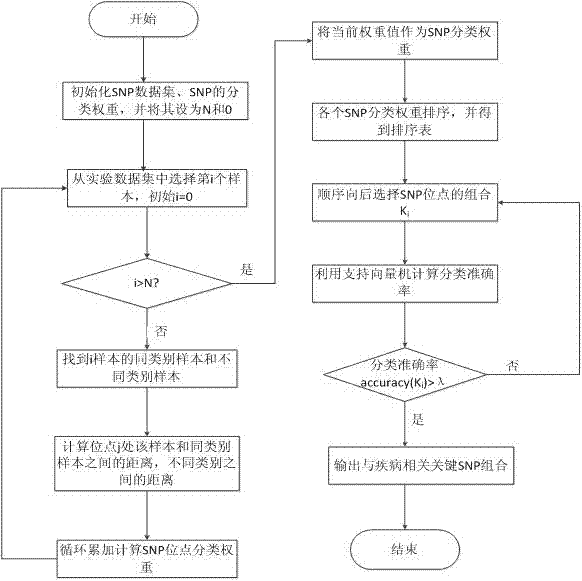
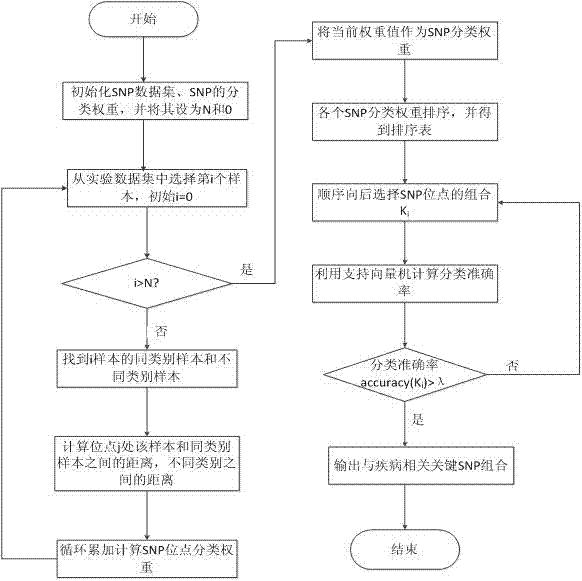
[0028] refer to figure 1 , a screening method for SNP data, the specific steps are as follows:
[0029] 1. Using the effect of a single SNP and the interaction between SNPs to calculate the SNP classification weight;
[0030] (1) Initialize the SNP data set and the classification weight of the SNP, and set them to N and 0 respectively;
[0031] (2) Select the i-th sample from the experimental data set N, if i>N, it will end, and take the current classification weight value as the final weight value, otherwise continue;
[0032] (3) Find the nearest neighbor sample m of the same category and the nearest neighbor sample n of different categories of the i sample, and the sample category only has two states: normal and diseased;
[0033] (4) Calculate the distance between the selected sample i at the location j and the nearest neighb...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com