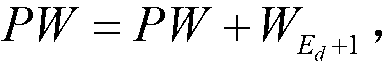Molecular dynamics accelerating method based on CUP (Central Processing Unit) and GPU (Graphics Processing Unit) cooperation
A molecular dynamics and molecular technology, applied in the field of molecular dynamics acceleration based on CPU and GPU collaboration, can solve problems such as low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0043] specific implementation plan
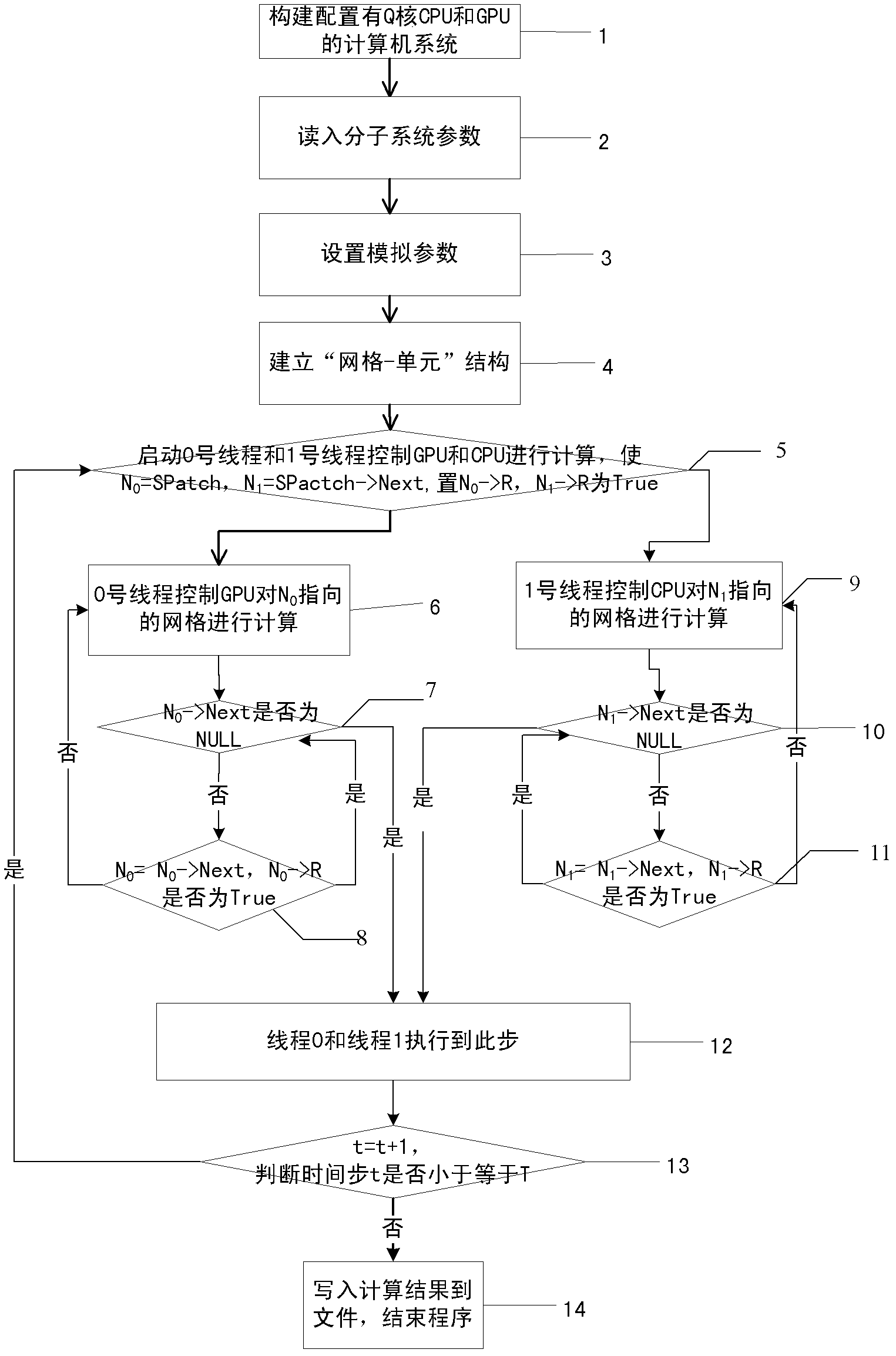
[0044] figure 1 Is the general flowchart of the present invention.
[0045] Step 1), building a computer system configured with Q core CPU and GPU;
[0046] Step 2), read in the molecular system parameters from the hard disk including the truncation radius R c and molecular data volume;
[0047] Step 3), setting the simulation parameters including the target grid weight TW, the target simulation step number T and the actual simulation step number t, and initializing the actual simulation step number t as 0.
[0048] Step 4), establish a "grid-unit" structure for the target molecular system, including the structure array SCell and the linked list SPatch;
[0049] Step 5), start No. 0 thread and No. 1 thread control GPU and CPU to calculate, will N 0 Point to SPatch, put N 1 Point to SPatch→Next, set N 0 → R is True, set N 1 → R is True, thread No. 0 goes to step 6), thread No. 1 goes to step 9);
[0050] Step 6), No. 0 thread contr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com