Method of increasing differentiation of chondrogenic progenitor cells
A technology of progenitor cells and cartilage, applied in the field of peptides of chondrogenesis, can solve the problem of inconvenient interpretation of cartilage differentiation variability and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
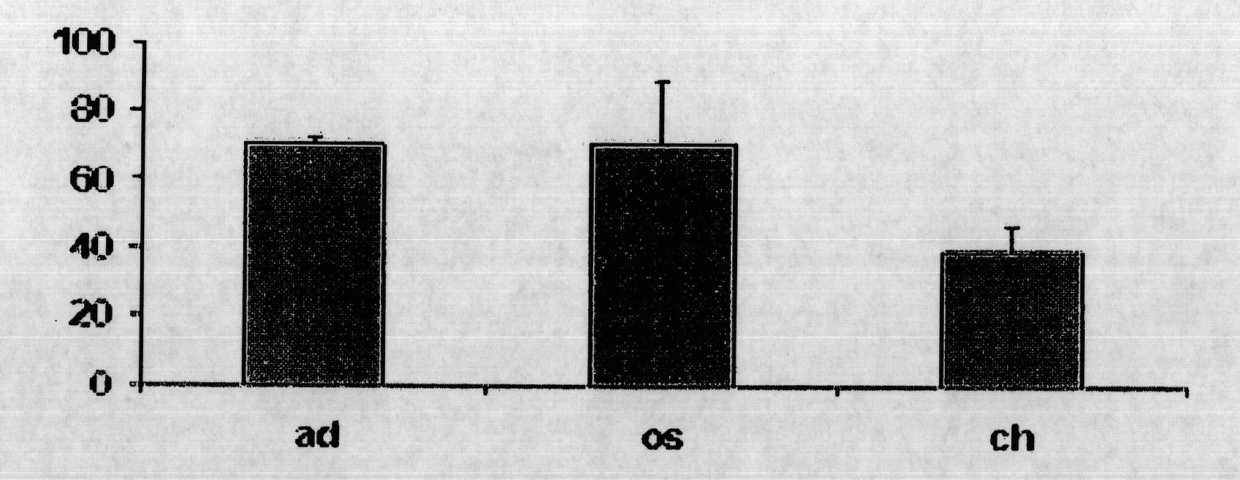
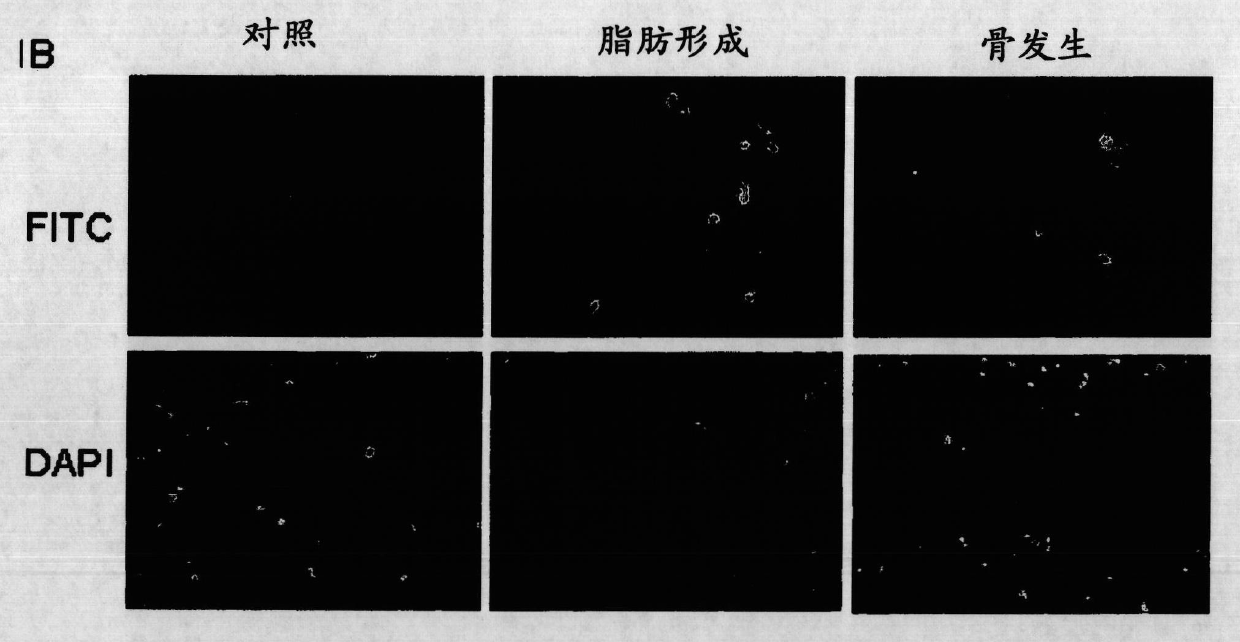
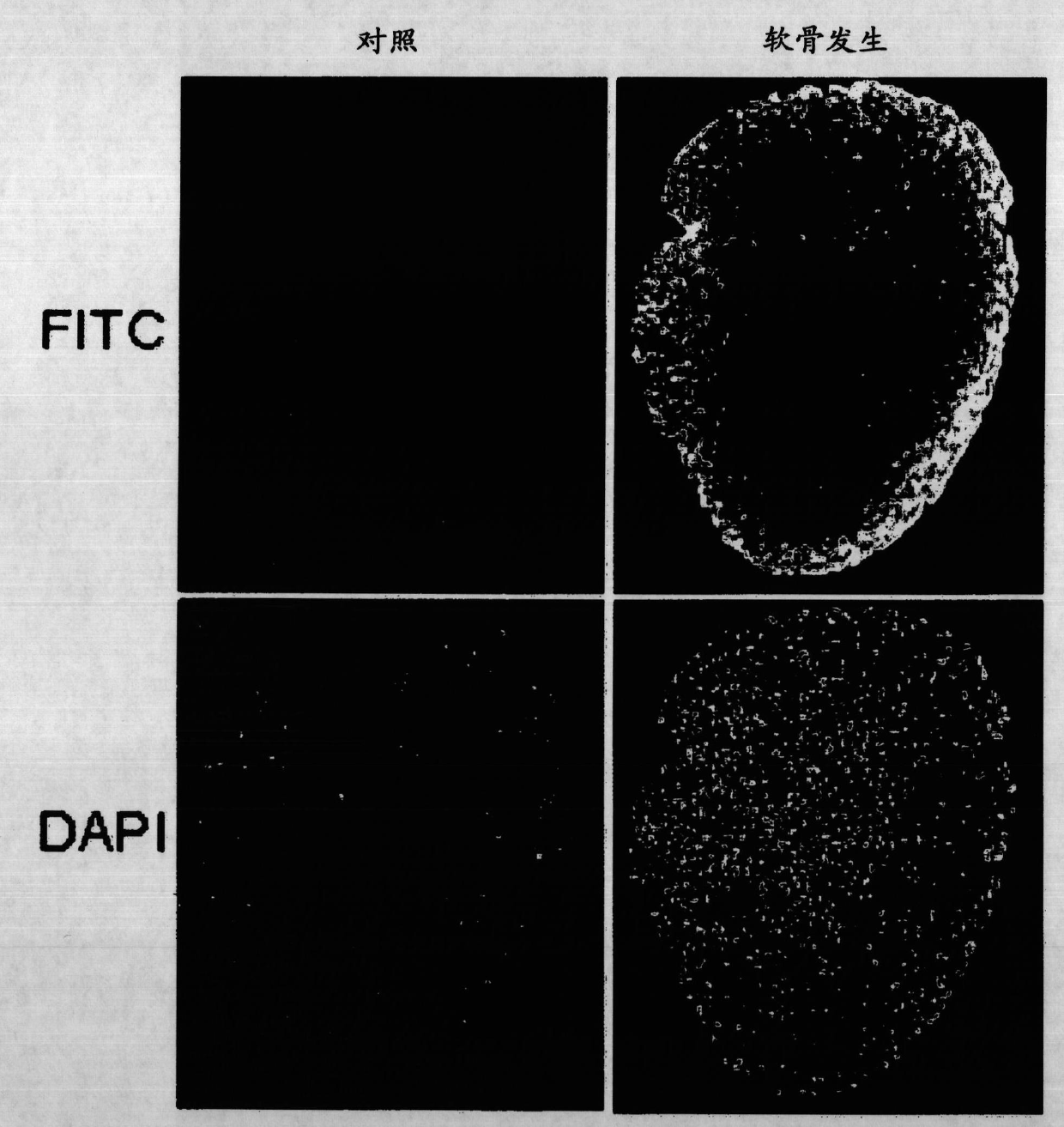
[0459] Example 1. MSC cultures and differentiation of osteoblasts, chondrocytes and adipocytes
[0460] After informed consent, human bone marrow-derived mesenchymal stem cells (hBMSCs) were collected from the iliac crest as described (Sekiya et al., 2002) according to the guidelines of the National University Hospital of Singapore and cultured. to cultivate.
[0461] To prevent spontaneous differentiation, cells were maintained at subconfluent levels. MSCs were induced to differentiate into adipocytes and osteoblasts as described in (Liu et al., 2007), and 2×10 5 and 1.5×10 5 MSCs were induced to differentiate into adipocytes and osteoblasts in adipogenic and osteogenic media, respectively, for 14 days.
[0462] Adipogenic medium contained 0.5 mM isobutyl-methylxanthine (IBMX), 1 μM dexamethasone (Sigma), 10 μM insulin, 200 μM indomethacin and 1% antibiotic / antimycotic. Osteogenic medium contained 0.1 μΜ dexamethasone, 50 μΜ ascorbic acid-2-phosphate, 10 mM β-glycerophosp...
Embodiment 2
[0465] Example 2. Construction of expression plasmids and infection to MSCs
[0466] ZNF145 was amplified from cDNA of MSCs undergoing osteogenesis for 14 days and then cloned into pEntry3C (Invitrogen).
[0467] Sox9 ultimate ORF clone was from Invitrogen. The pLentiviral vectors for overexpression of ZNF145 or Sox9 were generated by LR recombination between pEntry3C and pLenti6 / V5 (Invitrogen). The lentivirus was produced by co-transfecting the pLentiviral vector and packaging mix (Invitrogen) used to overexpress ZNF145 or Sox9 into 293FT cells, and then infected MSCs with the virus supernatant to achieve ZNF145 or Sox9 overexpression, and the resulting The MSCs were selected with 5 μg / ml blasticidin for 7 days. Empty pLenti6 / V5 without insert was used as control (empty).
Embodiment 3
[0468] Example 3. Quantitative real-time PCR
[0469]To quantify the effect of ZNF145 overexpression or knockdown on MSC differentiation, quantitative real-time PCR was performed with a Taqman expression assay (as described by the manufacturer) and ABI 7700 Prism (Applied Biosystems).
[0470] Briefly, 0.3 μg of total RNA was converted into 30 μl of cDNA using the Large Capacity cDNA Library Kit, which was then diluted to 300 μl. Quantitative RT-PCR was performed as follows: initial denaturation at 50 °C for 2 min, 95 °C for 10 min, followed by 40 PCR cycles (95 15 seconds at 60°C, 1 minute at 60°C).
[0471] All probes were designed with 5' fluorescence generating 6-carboxyfluorescein and 3' quencher - tetramethyl-6-carboxyrhodamine. Expression of human GAPDH was used to normalize gene expression levels.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com