Reference molecule for specifically detecting genetically modified rapeseed RT73 and application of reference molecule
A transgenic, rapeseed technology, applied in the biological field, can solve problems such as affecting the stability of standard molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] Embodiment 1, standard molecular construction
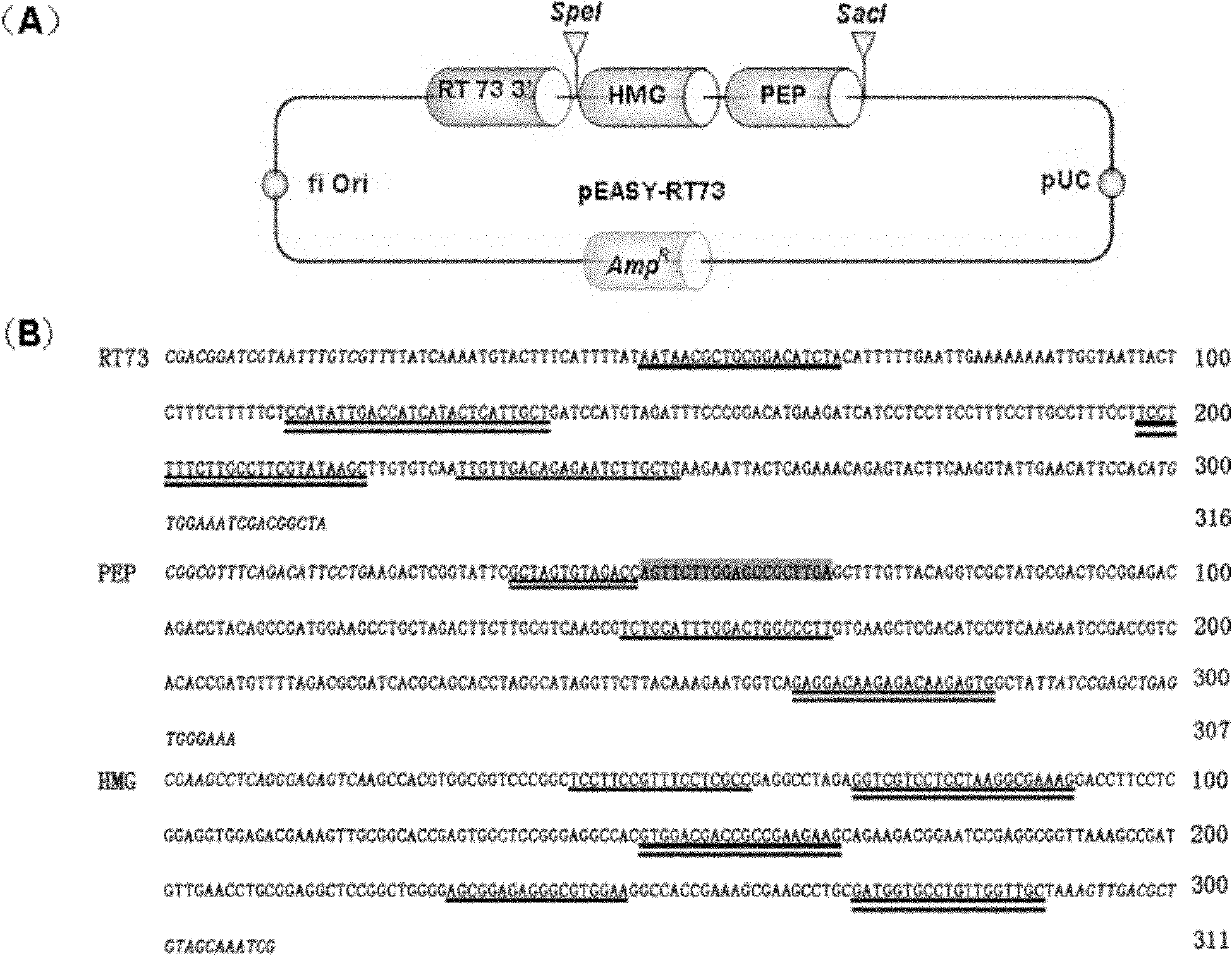
[0095] In order to construct a standard molecule suitable for the specific detection of transgenic rapeseed RT73 strains, the RT73' end adjacent sequence (316bp), rapeseed endogenous standard gene PEP (307bp) and HMG (311bp) three fragments were cloned into the pEASY-T3 vector, Get pEASY-RT73 ( figure 1 ), as a standard molecule, the size of the standard molecule is 3935bp. The results of three times of plasmid whole gene sequencing were consistent with the target sequence, indicating that the plasmid pEASY-RT73 was successfully constructed.
Embodiment 2
[0096] Embodiment 2, standard molecule is used for the applicability identification of ordinary PCR detection
[0097] (a) Specificity test
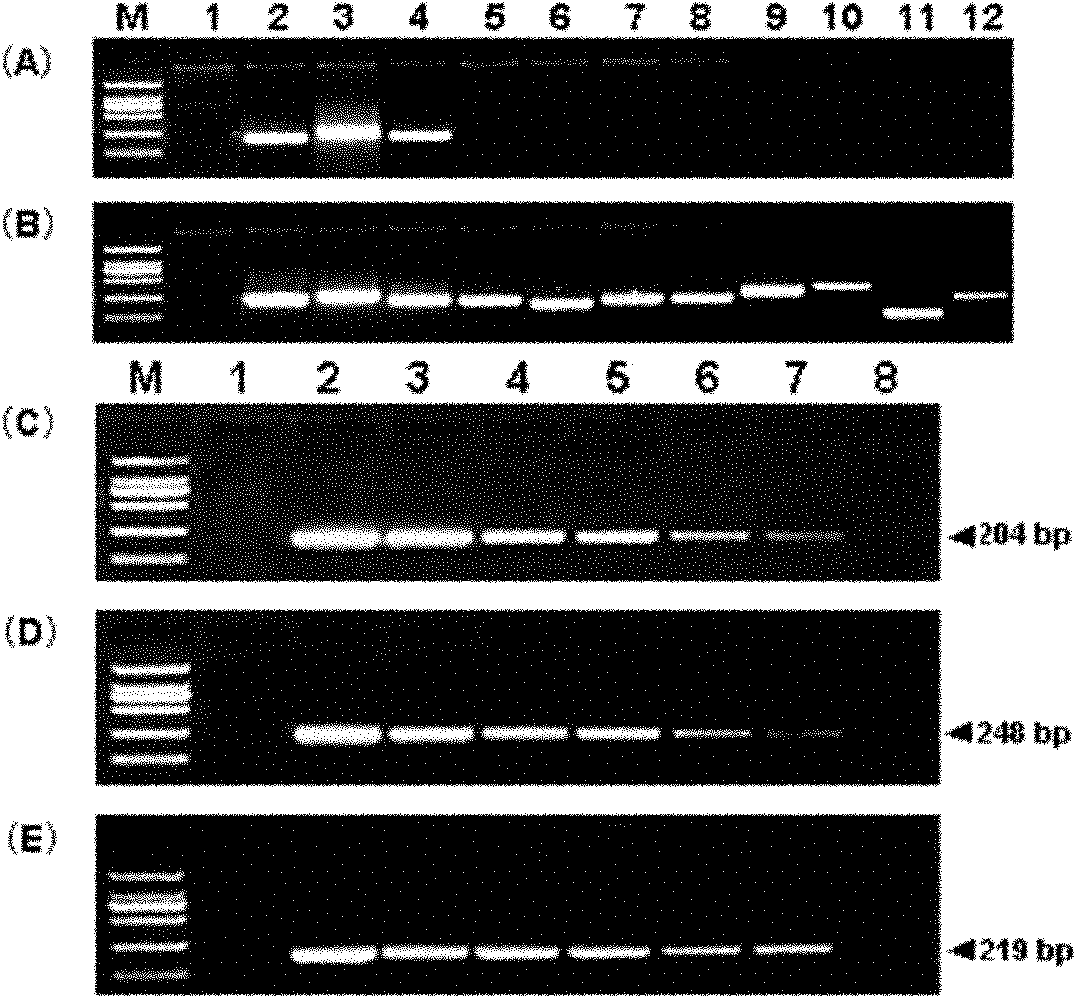
[0098] Nine kinds of transgenic rape strain-specific detection primers were used to test the specificity of standard molecules, and the results were as follows: figure 2 shown. In the amplification using standard molecular DNA as a template, obvious target bands can only be obtained in the amplification of the strain-specific sequence at the 3' end of RT73' and endogenous standard genes PEP and HMG ( figure 2 A).
[0099] There were no visible bands when amplified with transgenic rapeseed T45, Oxy235, MS8, MS1, RF1, RF2, RF3 and Topas19 / 2 strain-specific detection primers, but the positive controls corresponding to each detection system had target bands ( figure 2 B).
[0100] Therefore, the standard molecule pEASY-RT73 is highly specific for the detection of the transgenic rapeseed RT73 line.
[0101] (b) Detection sensitivity ...
Embodiment 3
[0103] Embodiment 3, standard molecule is used for the applicability identification of real-time fluorescent PCR detection
[0104] (a) Preparation of standard curve
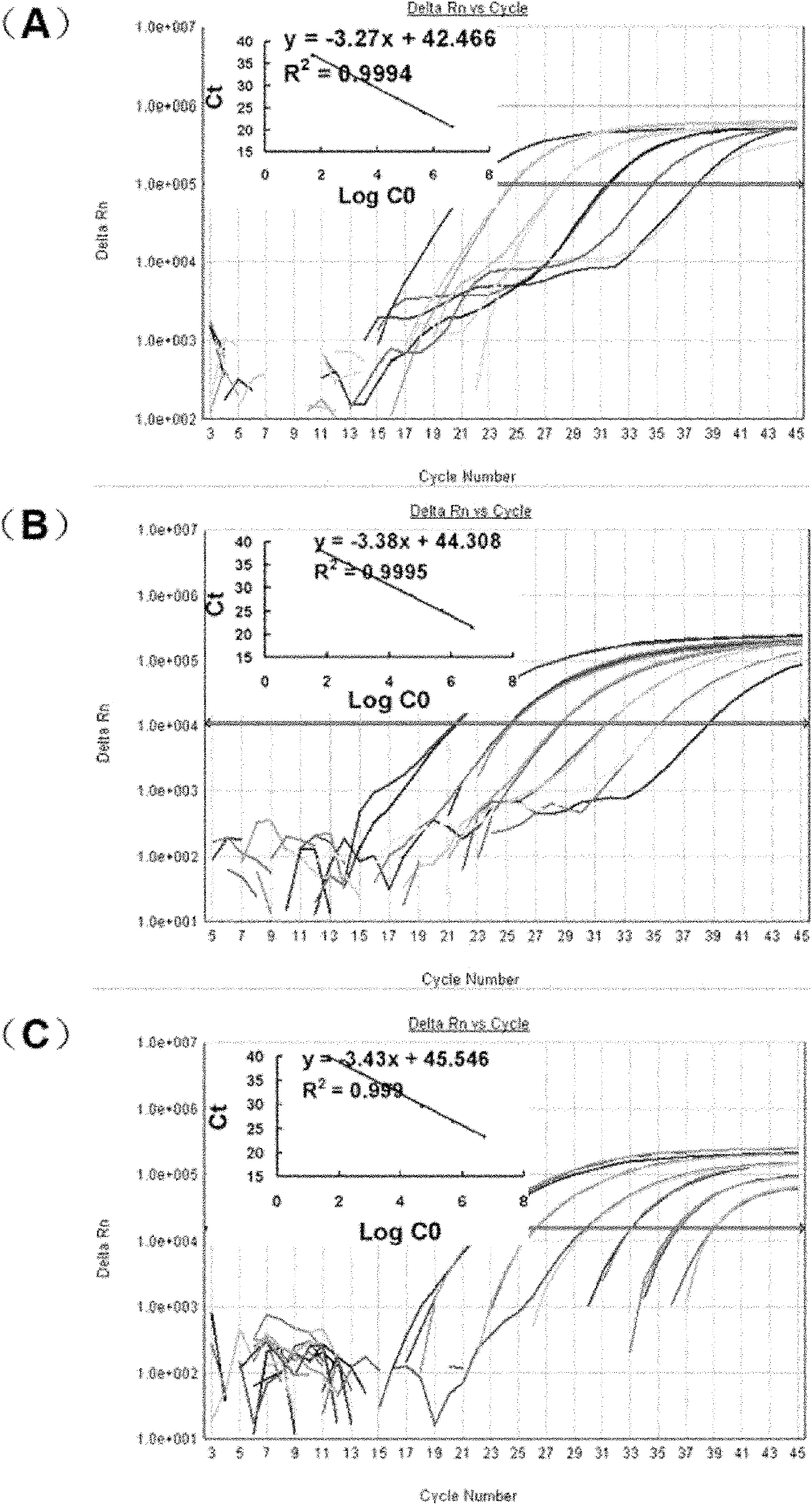
[0105] Using gradiently diluted standard molecular DNA as a template, real-time fluorescent PCR amplification was performed on three target fragments (RT733' adjacent sequence, HMG, and PEP) to make a standard curve. like image 3 As shown, the slopes of the standard curves of the three target fragments were -3.27, -3.38 and -3.43, respectively, and the reaction efficiencies were 1.02, 0.98 and 0.96, R 2 All greater than 0.999.
[0106] Therefore, the standard curve is suitable for further quantitative analysis of the samples.
[0107] (b) Limit of detection (LOD) and limit of quantitation (LOQ) tests
[0108] Among the 25 amplifications using 50 copies of DNA as a template, RT73 strain-specific sequence, PEP and HMG three target fragments had 25 amplification curves, and the RSD values were 2.04%, 3.06% a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com