Method for screening compound with targeted action on inactive conformation of protein kinase
A technology of inactive conformation and protein kinase, which is applied in the field of protein structure prediction and virtual screening of drug molecules, and can solve problems such as long-term treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0079] The method of the present invention is applied to the calculation process of Mycobacterium tuberculosis protein kinase PknB:
[0080] The first step, the construction of the kinase inactive conformation ensemble:
[0081] a) Download the PDB file 1O6Y of PknB protein kinase, remove the heteroatoms in it, and perform preliminary optimization after filling;
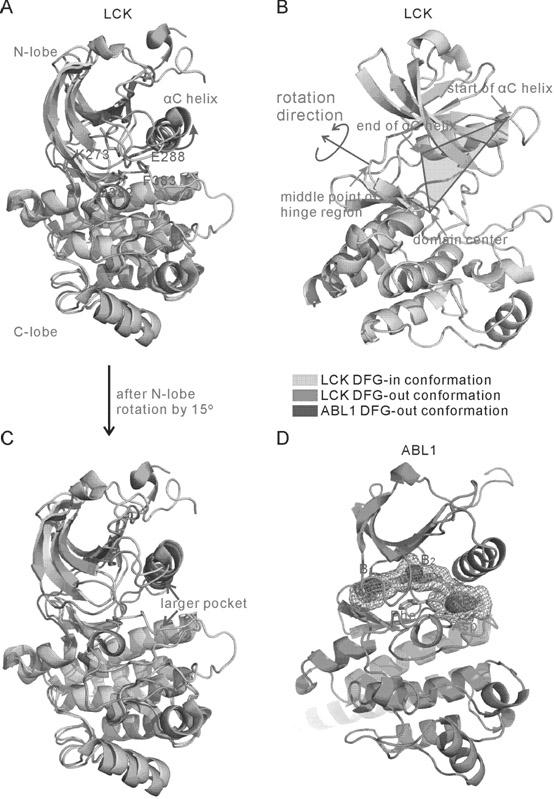
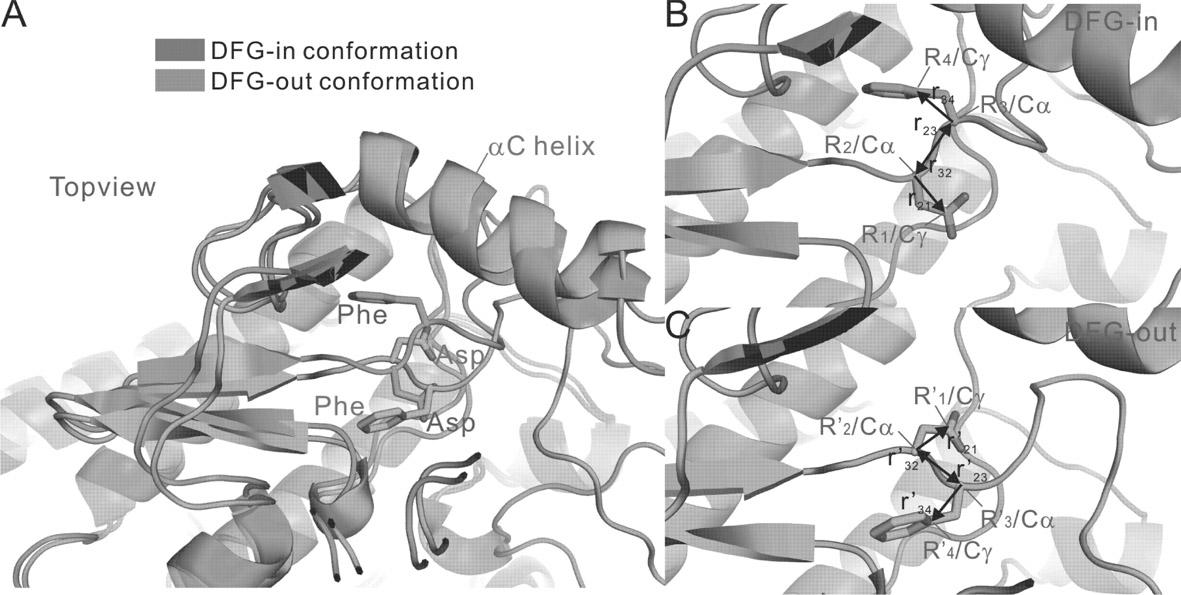
[0082] b) As mentioned above, extract the coordinates of 4 atoms to calculate the sum of the required distances, and the value is 29.64 ?, indicating that the N-lobe of 1O6Y in front of the active chain segment needs to be rotated outward by 15 degrees;
[0083] c) Obtain the desired initial conformation after rotation, and calculate the remodeling of the active segment after setting each parameter file;
[0084] d) Among the 200 prediction models obtained, 163 were judged as DFG-out conformation by vector method;
[0085] e) These conformations are tested for the space size of the active site, and 8 conformation...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com