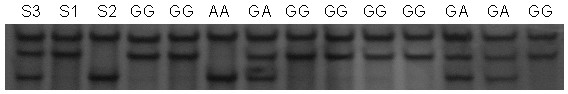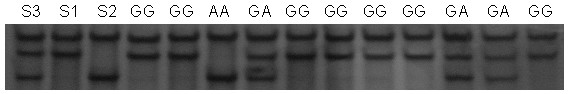Method and kit for detecting correlated mutation allele of total farrow number of sow as well as application
A technology of total litter size and alleles, applied in the field of alleles, can solve the problems of lack of correspondence, low selection efficiency, and inability to directly know individual genotypes, etc., to achieve economic benefits, high application value, and detection costs cheap effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Embodiment 1: the selection of excellent breeding pigs in the breeding of pigs
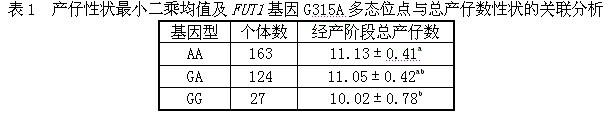
[0020] 1. Establishment of PCR-SSCP detection method and determination of polymorphic sites
[0021] 1. PCR amplification
[0022] Primers were designed according to the porcine GenBank Accession Number AX752829 sequence, and the primers were as follows:
[0023] U (upstream primer): 5'-ATCTCCCGTGGCCATATTCT-3' (SEQ ID NO: 1);
[0024] D (downstream primer): 5'-ATGGCAGGCTGGATGAAG-3' (SEQ ID NO: 2).
[0025] The Beijing black pig was selected as the experimental material. Using the genomic DNA of Beijing black pig as a template, PCR amplification was performed using primers U and D.
[0026] Amplification system: Genomic DNA 200ng, 10×PCR amplification buffer 2.5μl, dNTPs 5mM, upstream and downstream primers 50ng each, Taq DNA polymerase 0.75U, Mg 2+ 2.5mmol / L, with ddH 2 O Supplement the reaction system to 25 μl.
[0027] The PCR reaction conditions were: denaturation at 95°C for 5 min...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com