Method for obtaining nucleotide sequence in V3 region of 16S rRNA genes of bacteria and special primer thereof
A nucleotide sequence, nucleotide technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of long test period, cumbersome test steps, heavy workload, etc. Experiment cost, long experiment period, and the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1, discovery and analysis of specific primer pair
[0035] 1. Discovery of specific primer pairs
[0036] DNA fragment A (16S rRNA gene of Clostridium bifermentans strain MKA2) shown in sequence 1 of the sequence listing was synthesized. The DNA shown in Sequence 1 is complementary to the DNA shown in GENBANK ACCESSION NUMBER GU358061.1. The V3 region of the 16S rRNA gene of Clostridium bifermentans strain MKA 2 is from the 125th to the 299th nucleotide at the 5' end.
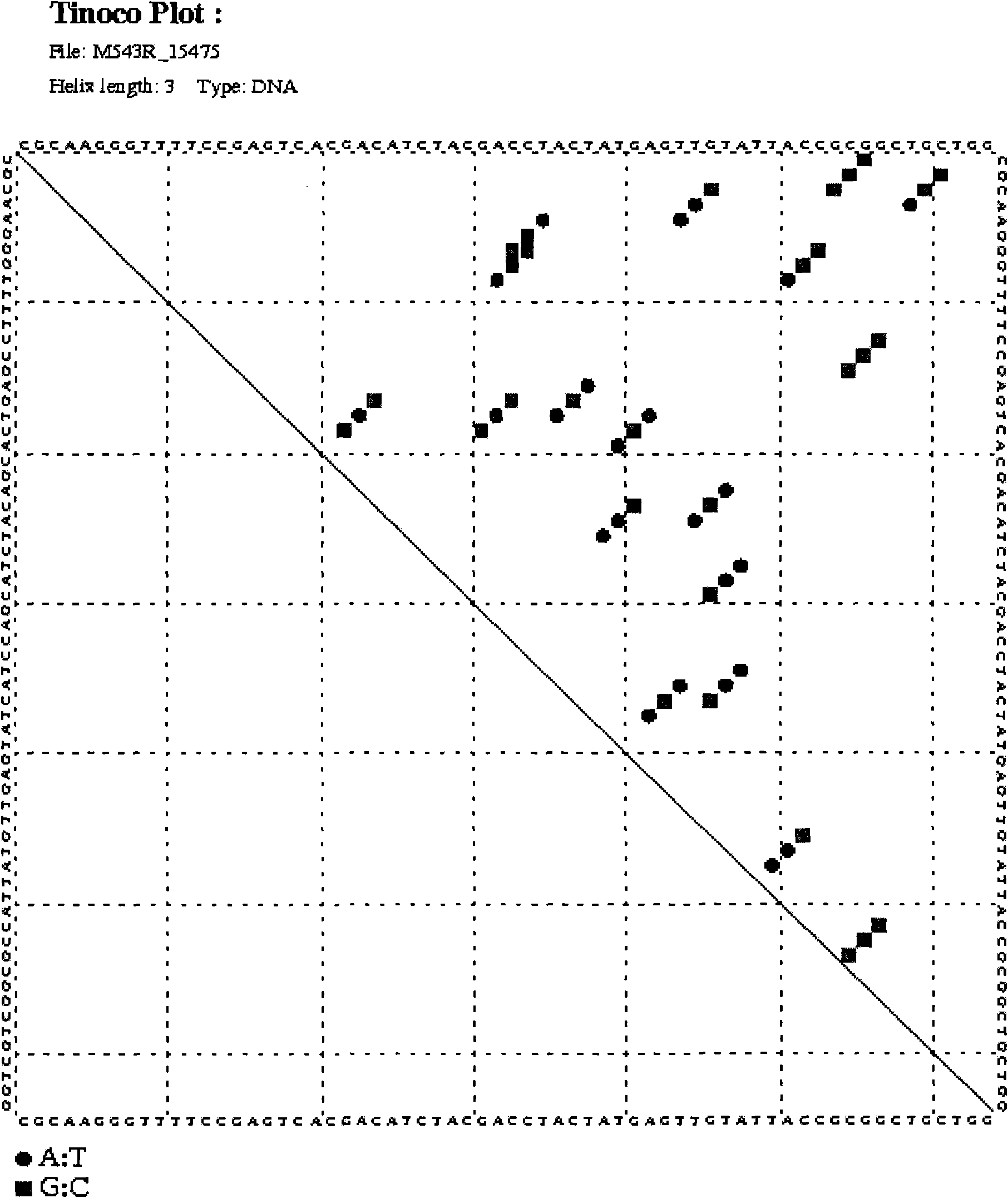
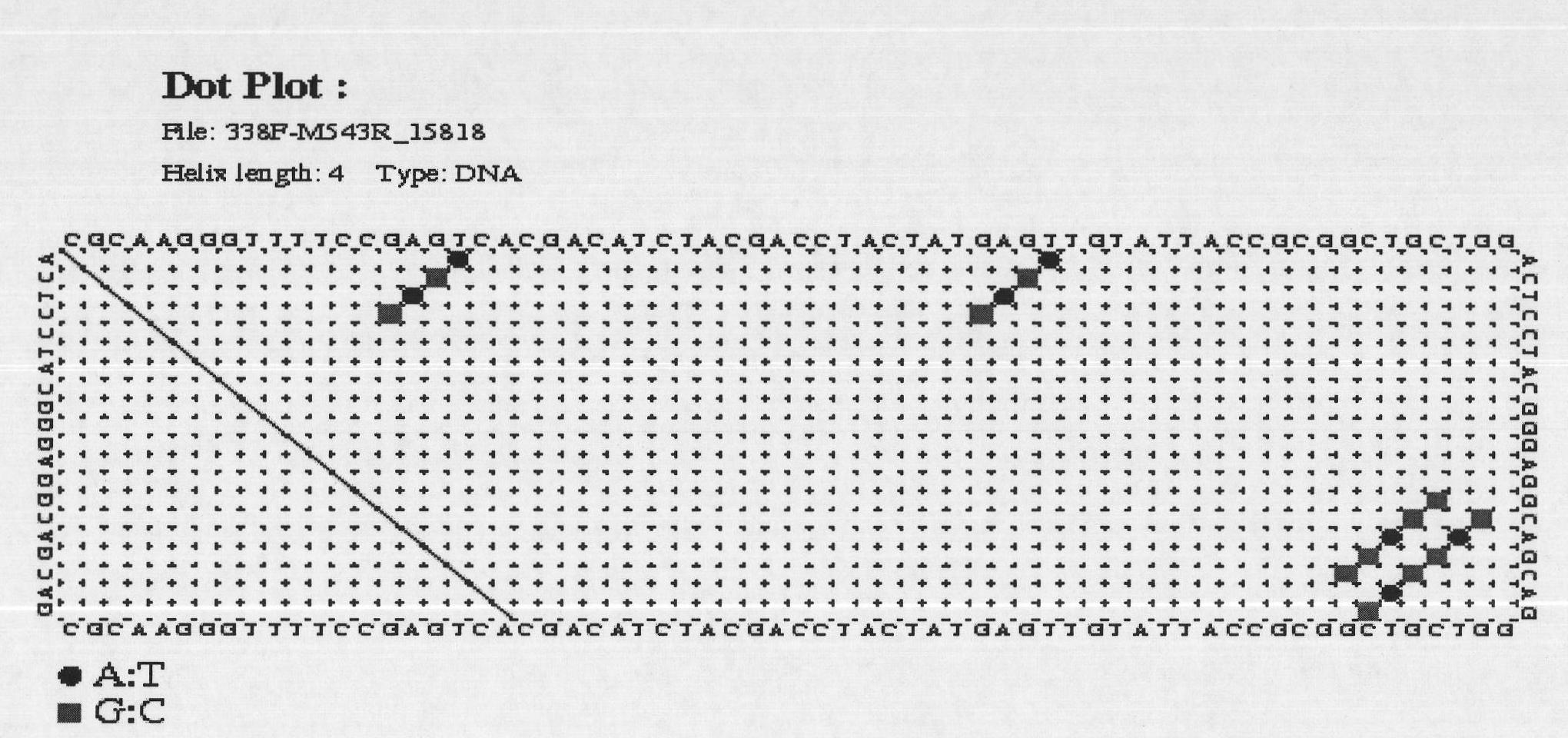
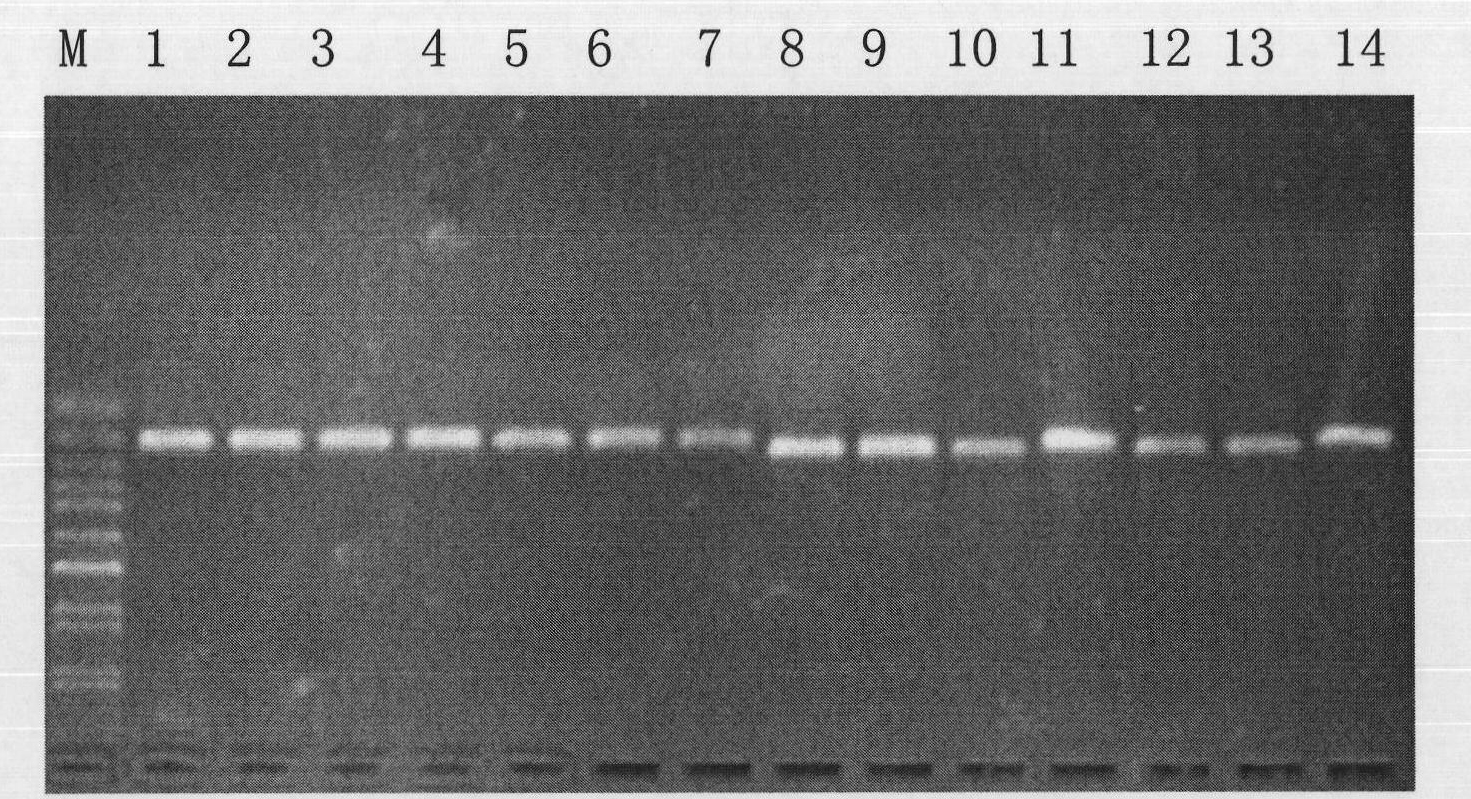
[0037] DNA fragment A was amplified by PCR with universal primers (338F-543R). The PCR product was subjected to 1.5% (mass percentage) agarose gel electrophoresis, and then purified by cutting gel with Tiangen DNA gel purification kit. The elution volume of the purified product was 50ul. The purified DNA was directly sequenced using 543R as a sequencing primer. Sequence map see Figure 4 A, for the sequencing results, see sequence 2 in the sequence listing. The analysis found that: when...
Embodiment 2
[0046] Embodiment 2, the acquisition of the V3 region nucleotide sequence of bacterial 16S rRNA gene
[0047] DNA fragment B (16S rRNA gene of Shewanella sp. enrichment culture clone LY4) shown in sequence 5 of the sequence listing was synthesized. The DNA shown in Sequence 5 is complementary to the DNA shown in GENBANK ACCESSION NUMBER GQ465946.1. The V3 region of the 16S rRNA gene of Shewanella sp.enrichment culture clone LY4 is from the 195th to the 395th nucleotide at the 5' end.
[0048] 1. Use universal primers (338F-543R) to amplify DNA fragment B by PCR. The PCR product was subjected to 1.5% (mass percentage) agarose gel electrophoresis, and then purified by cutting gel with Tiangen DNA gel purification kit. The elution volume of the purified product was 50ul. The purified DNA was directly sequenced using 543R as a sequencing primer. Sequence map see Figure 6 A, for the sequencing results, see sequence 6 in the sequence listing.
[0049] 2. Use the specific prim...
Embodiment 3
[0052] Embodiment 3, the acquisition of the V3 region nucleotide sequence of bacterial 16S rRNA gene
[0053] The DNA fragment C (the 16S rRNA gene of Marine bacterium SIMO-4432) shown in the sequence 9 of the sequence listing was synthesized. The DNA shown in Sequence 9 is complementary to the DNA shown in GENBANK ACCESSION NUMBER DQ469762.1. The V3 region of the 16S rRNA gene of Marinebacterium SIMO-4432 is from the 129th to the 327th nucleotide at the 5' end.
[0054] 1. Carry out PCR amplification of DNA fragment C with universal primers (338F-543R). The PCR product was subjected to 1.5% (mass percentage) agarose gel electrophoresis, and then purified by cutting gel with Tiangen DNA gel purification kit. The elution volume of the purified product was 50ul. The purified DNA was directly sequenced using 543R as a sequencing primer. Sequence map see Figure 8 A, for the sequencing results, see sequence 10 in the sequence listing.
[0055] 2. Use the specific primer pair...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com