Clone method of SGAE label 3' end cDNA segment
A cloning method and fragment technology, applied in the field of cloning of SGAE-labeled 3'-end cDNA fragments, can solve the problems of cumbersome process, many non-specific bands, and large amount of initial RNA, and achieve simple process, low cost, and uniform length Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The embodiments of the present invention are described in detail below in conjunction with the accompanying drawings: this embodiment is implemented under the premise of the technical solution of the present invention, and detailed implementation methods and processes are provided, but the protection scope of the present invention is not limited to the following implementations example. The implementation method that does not indicate specific conditions in the following examples is usually according to conventional conditions, such as the molecular cloning of Sambrook et al.: the conditions described in the laboratory manual (New York: Cold Spring Harbor Laboratory Press, 1998), or according to the manufacturer's suggested conditions.
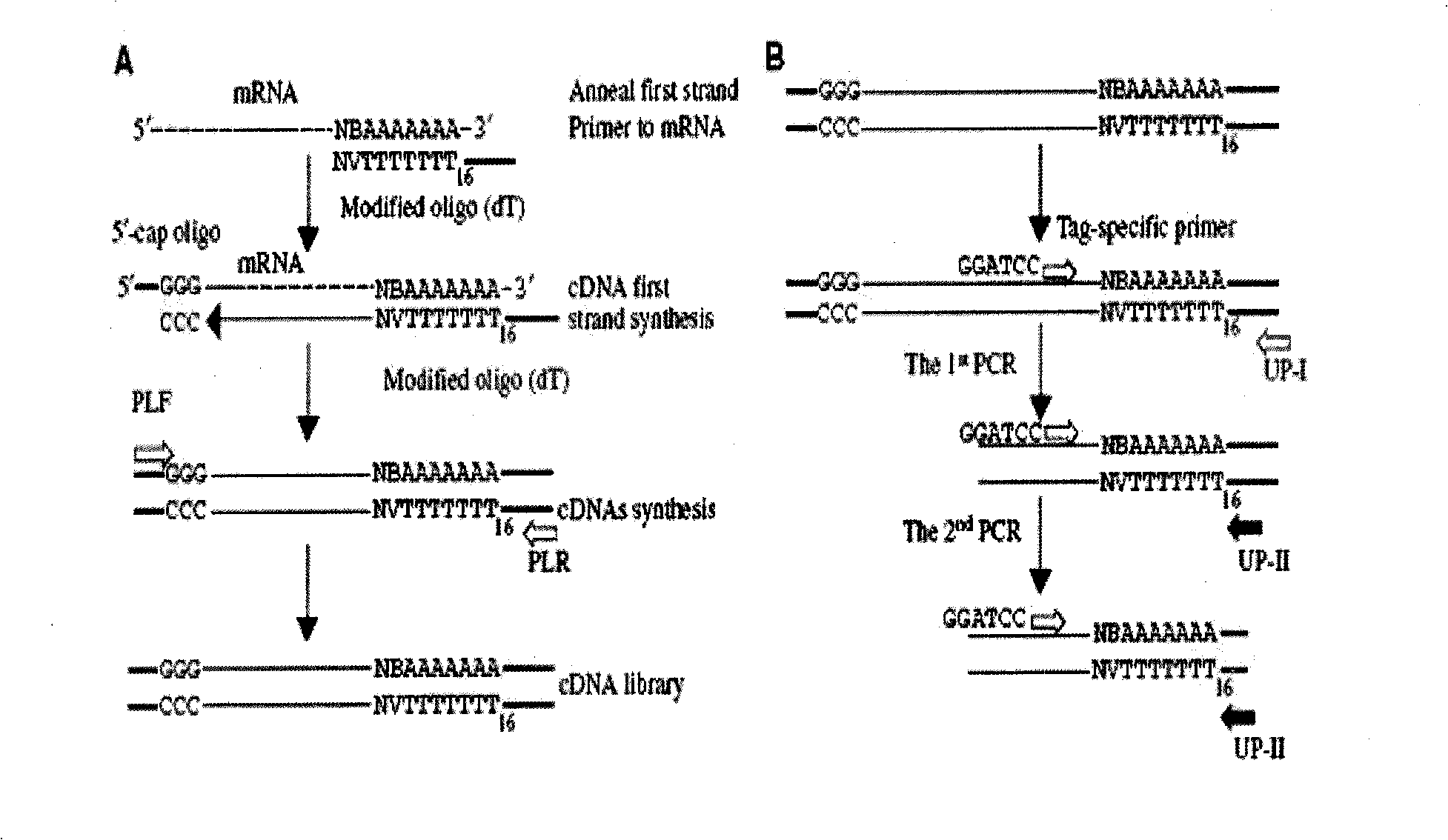
[0029] figure 1 For the explanation of total cDNAs amplification and 3' end cDNA fragmentation method (TAST-PCR) technical mechanism, among the figure, figure A is the amplification of cDNAs, at first use the anchor oligo(dT) that adds...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com