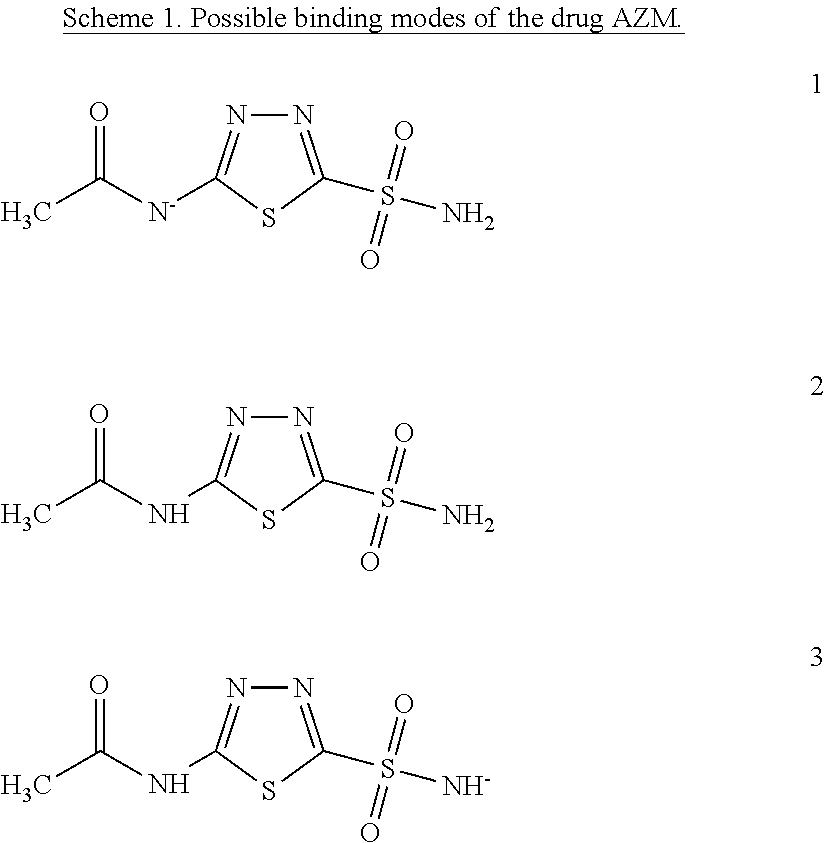
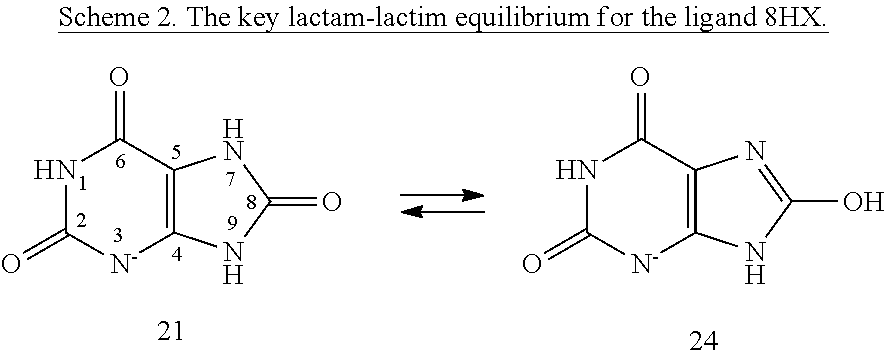
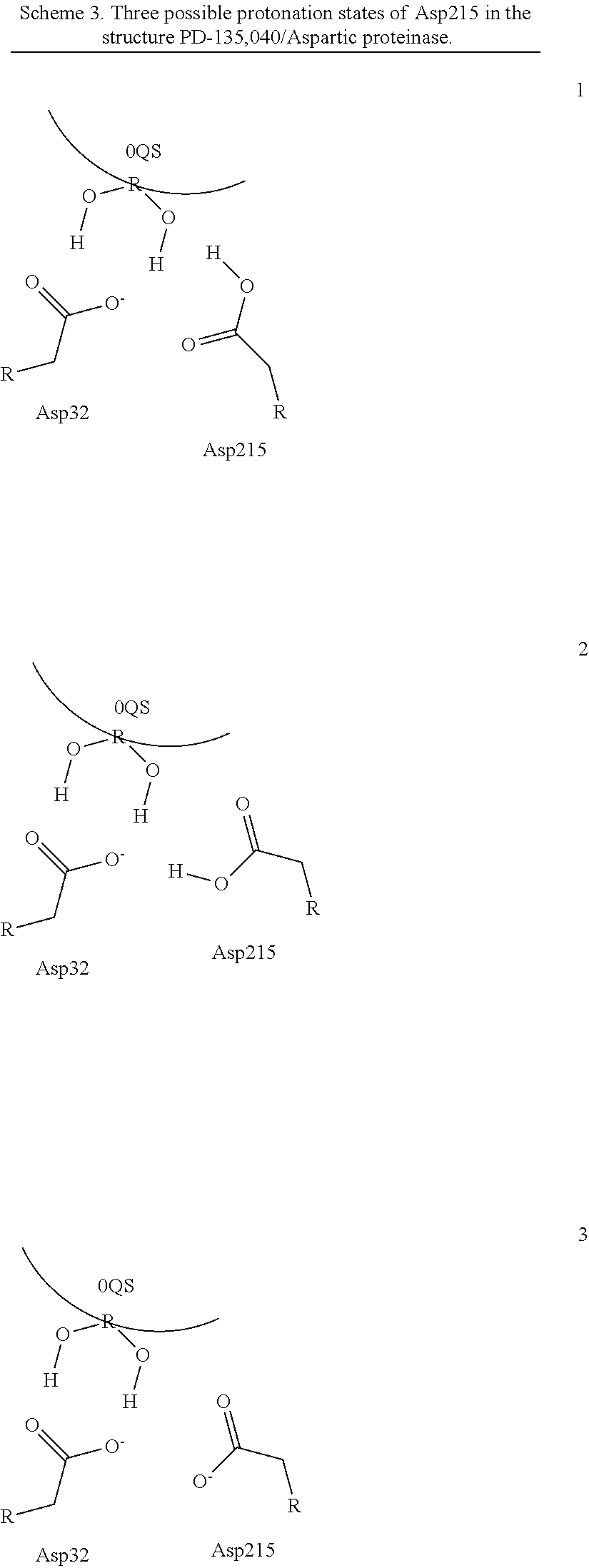
Quantum Mechanical/X-Ray Crystallography Diagnostic for Proteins
a technology of x-ray crystallography and quantum mechanical, applied in the direction of material analysis using wave/particle radiation, instruments, molecular structure, etc., can solve the problems of insufficient information about epitopes, protonation or solvent effects, x-ray crystallography, etc., to accurately assess protein conformation, protonation and solvent effects, useful and reliable
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
)
[0008]Within structure-guided drug discovery (SGDD) and structure-based drug discovery (SBDD), accurate understanding of the protein:ligand complex structure, including the relevant proper protonation, is crucial for obtaining meaningful results from docking / scoring, thermodynamic calculations, active site exploration, lead optimization, and ultimately, medicinal chemistry. The most ubiquitous element in the universe is hydrogen, and these protons are critical for exploring the chemistry within the active site. For example, in the drug Mirapex, which is used to treat symptoms of Parkinson's disease, the important chemical activity is conferred by a single aminothiazole tautomeric state rather than an alternative imino tautomer; thus the selection of the wrong state during the drug design would lead to irrelevant findings. This situation is not uncommon, and drug discovery frequently hinges on the determination of one state vs. another state. The present invention provides a verifia...
PUM
| Property | Measurement | Unit |
|---|---|---|
| quantum mechanical refinement | aaaaa | aaaaa |
| conformational | aaaaa | aaaaa |
| solvent effect | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com