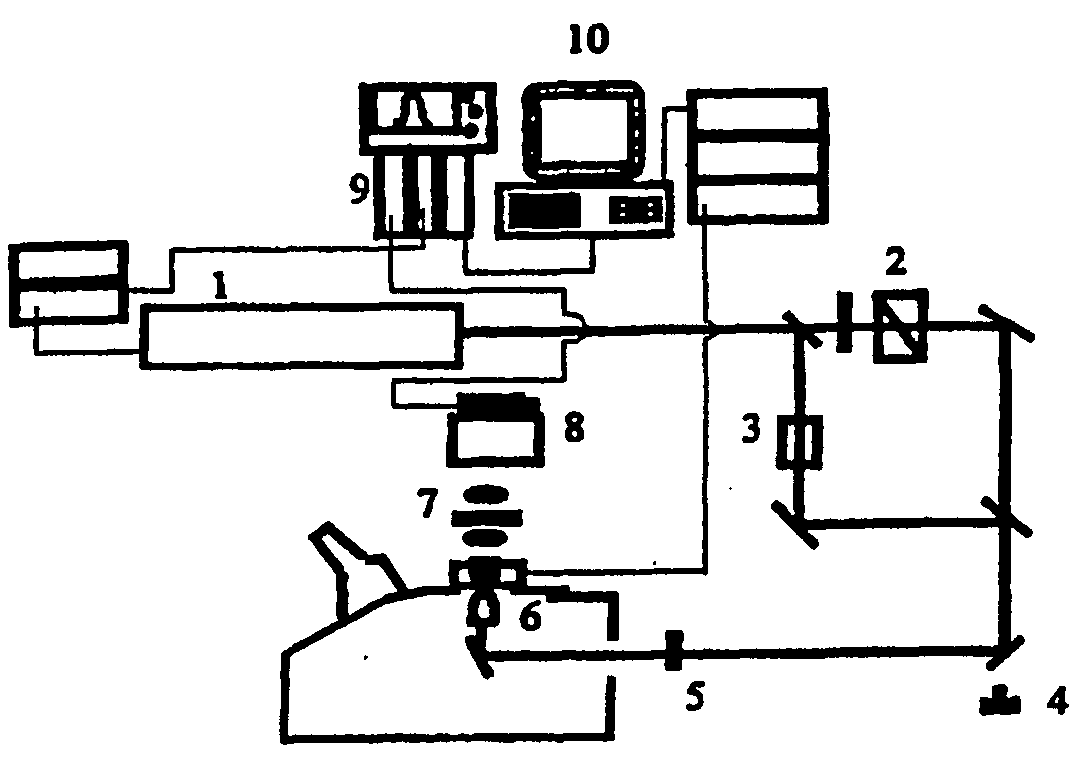
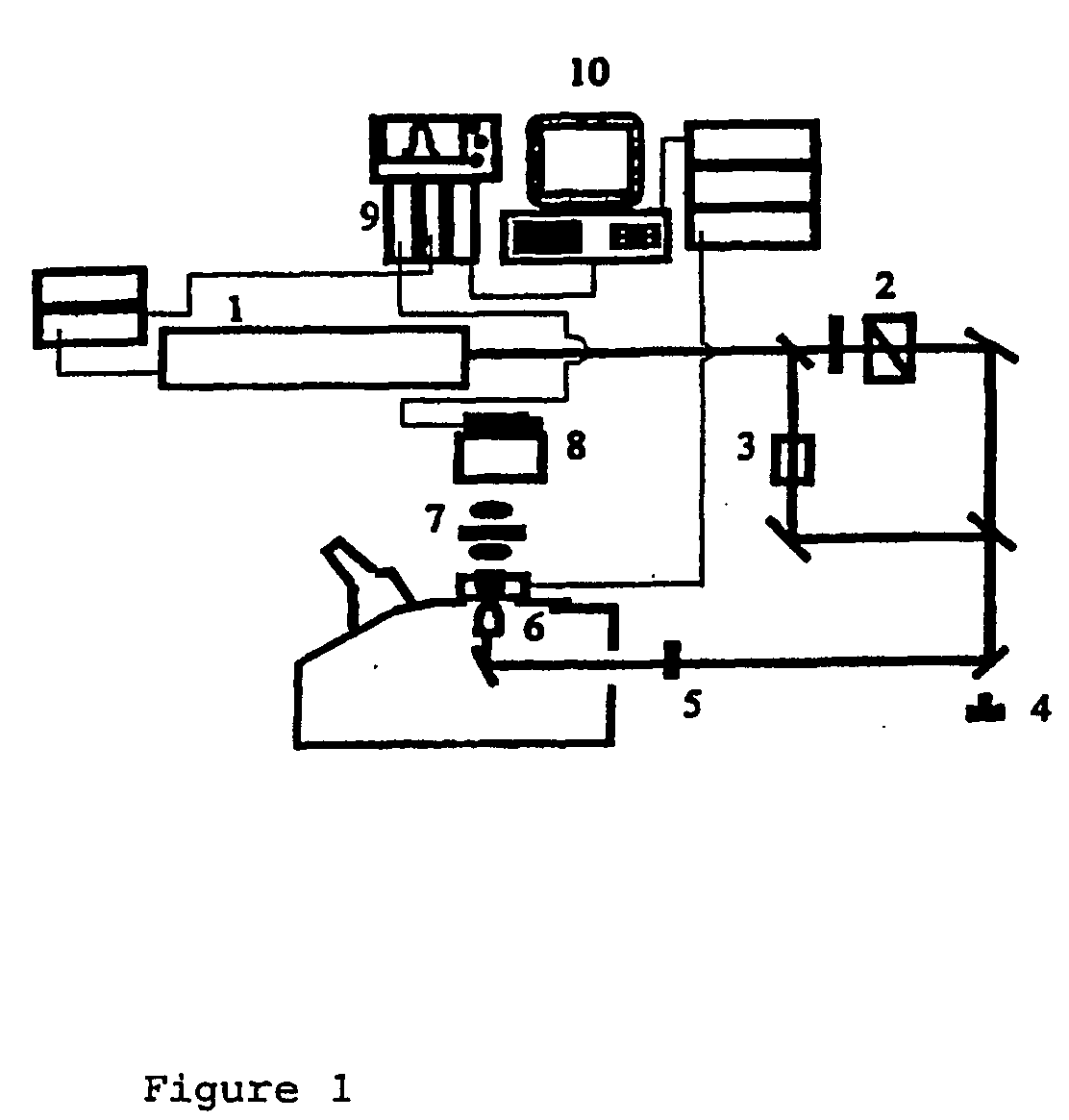
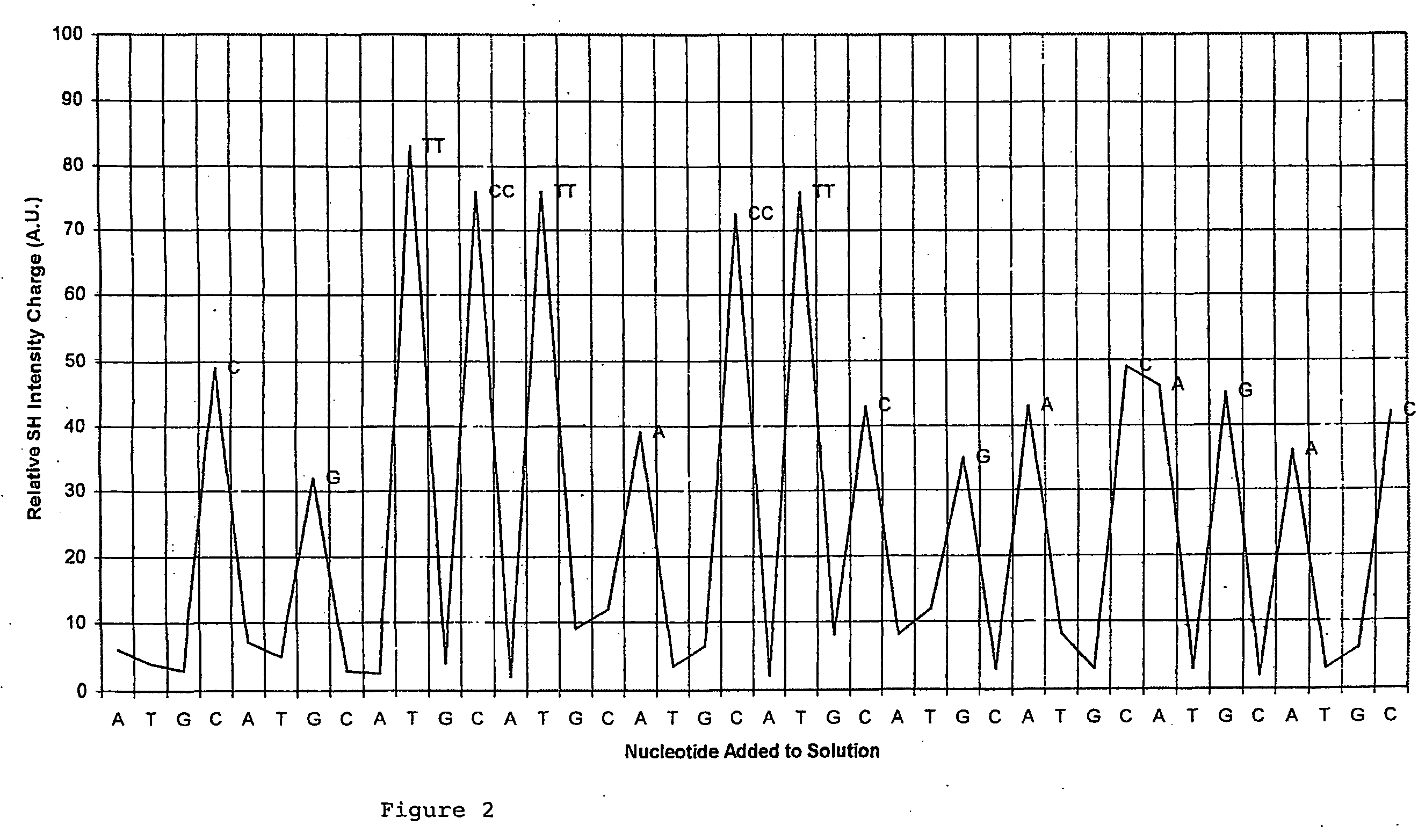
Polynucleotide Sequencing Method
a polynucleotide and sequencing method technology, applied in the field of polynucleotide sequence determination, can solve the problems of limiting the use of sequencing methods, increasing background interference from fluorophores, and slowness, and achieves reduced secondary structure considerations, extensive fragment reassembly, and long sequence read length.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018]The present invention makes use of conventional non-linear optical measurements to identify a conformational and / or mass and / or energy distribution change occurring as a polynucleotide processive enzyme interacts with the individual bases on a target polynucleotide or incorporates nucleotides onto a nascent polynucleotide molecule.
[0019]The use of non-linear optical methods for imaging molecules is known. What has not been appreciated is that these methods can be applied to the sequencing of a polynucleotide, making use of an immobilised or fixed enzyme.
[0020]In a separate embodiment, a linear signal is generated in addition to a non-linear signal and the linear signal is detected. The two signals are said to be coupled, resulting in enhanced detection.
[0021]The term “polynucleotide” as used herein is to be interpreted broadly, and includes DNA and RNA, including modified DNA and RNA, DNA / RNA hybrids, as well as other hybridising nucleic acid-like molecules, e.g. peptide nucle...
PUM
| Property | Measurement | Unit |
|---|---|---|
| enzyme activity | aaaaa | aaaaa |
| third harmonic generation imaging | aaaaa | aaaaa |
| optical detection | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com