Molecular docking technique for screening of combinatorial libraries
a combinatorial library and molecular docking technology, applied in the field of combinatorial library screening, can solve the problems that algorithms may encounter difficulties with compounds commonly found in combinatorial libraries, and achieve the effects of improving energy minimization algorithms, fast flexible docking methods, and removing significant burdens from conformational search procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
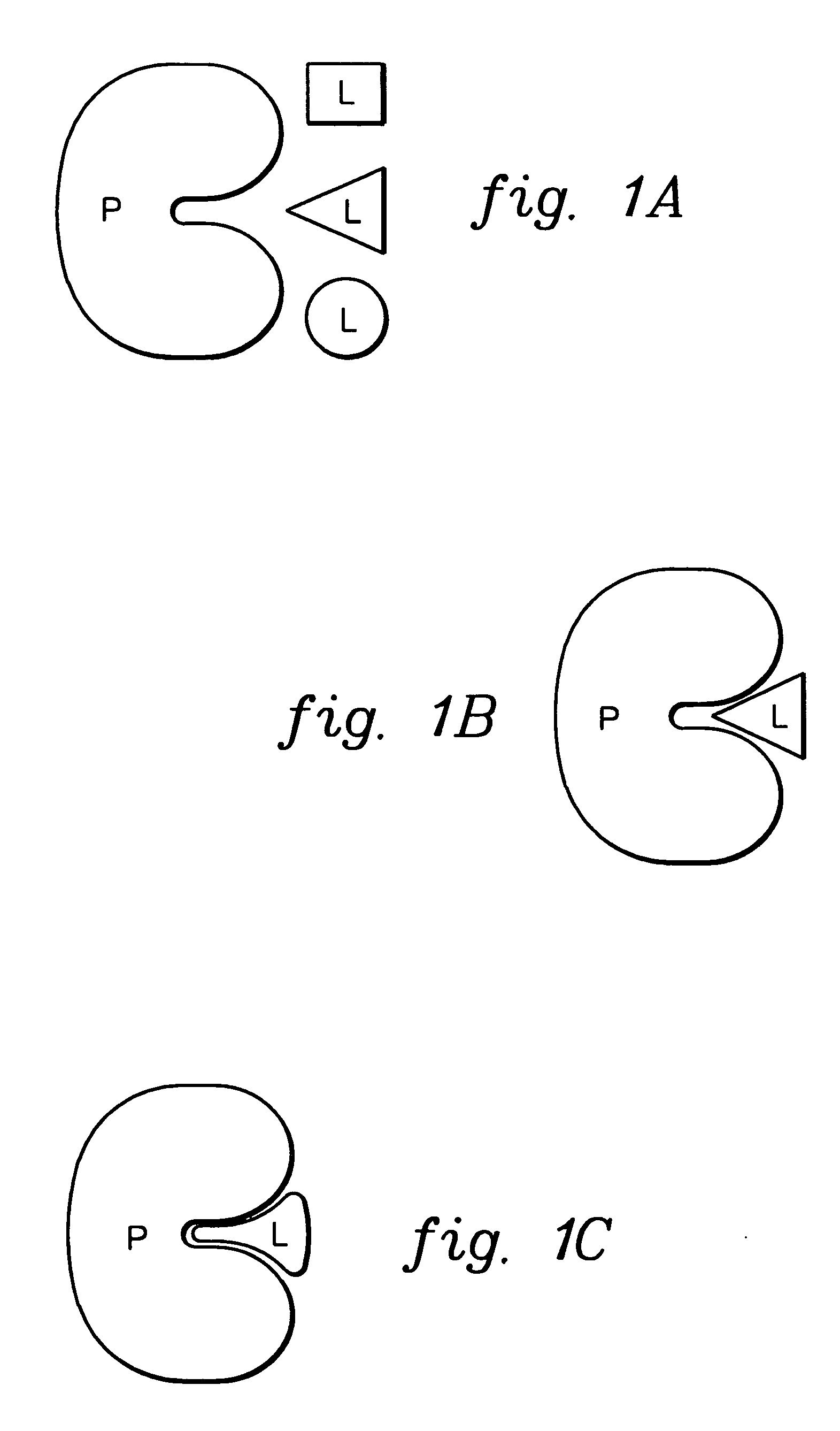
[0023] The docking procedure discussed below is based on a conceptual picture of protein-ligand complex formation (see FIGS. 1A-1C). Initially, the ligand (L) adopts many conformations in solution. The protein (P) recognizes one or several of these conformations. Upon recognition, the ligand, protein and solvent follow the local energy landscape to form the final complex.
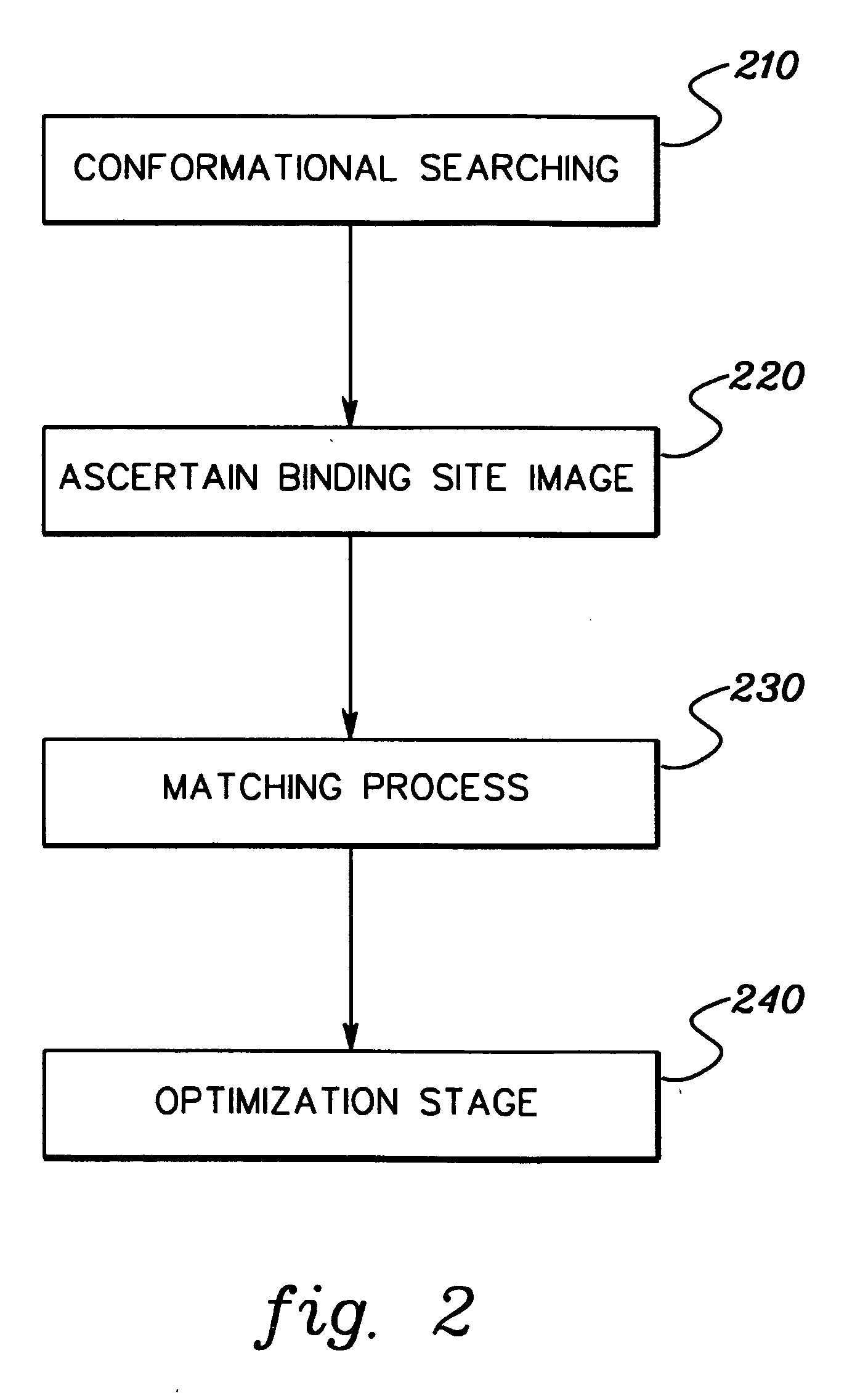
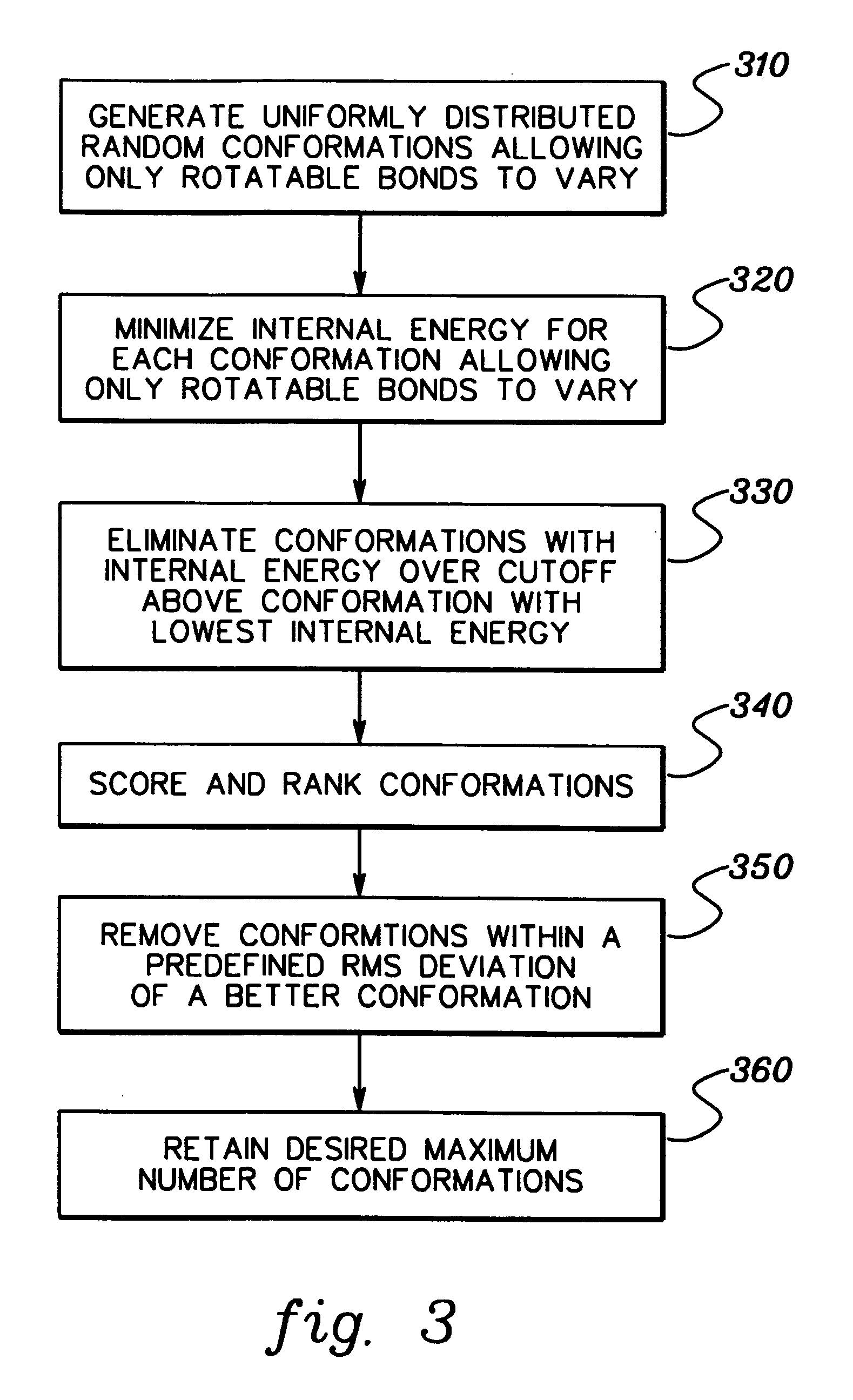
[0024] This simple picture of protein / ligand complex formation is converted into an efficient computational model in accordance with an aspect of the present invention, as follows. The initial solution conformations are generated using a straightforward conformational search procedure. One might view the conformational search part of this technique as part of the entire docking process, but since it involves only the ligand, it can be decoupled from the purely docking steps. This is justified since 3-D databases of conformations for a collection of molecules can readily be generated and stored for use in numerous d...
PUM
| Property | Measurement | Unit |
|---|---|---|
| internal energy | aaaaa | aaaaa |
| dihedral angles | aaaaa | aaaaa |
| dihedral angles | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com