Rolling circle amplification of RNA
a technology of rna and rna molecules, applied in the field of rna amplification, can solve problems such as amplification bias, and achieve the effect of increasing the amplification yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
A. Example 1
Circularization-dependent Amplification of Human RNA
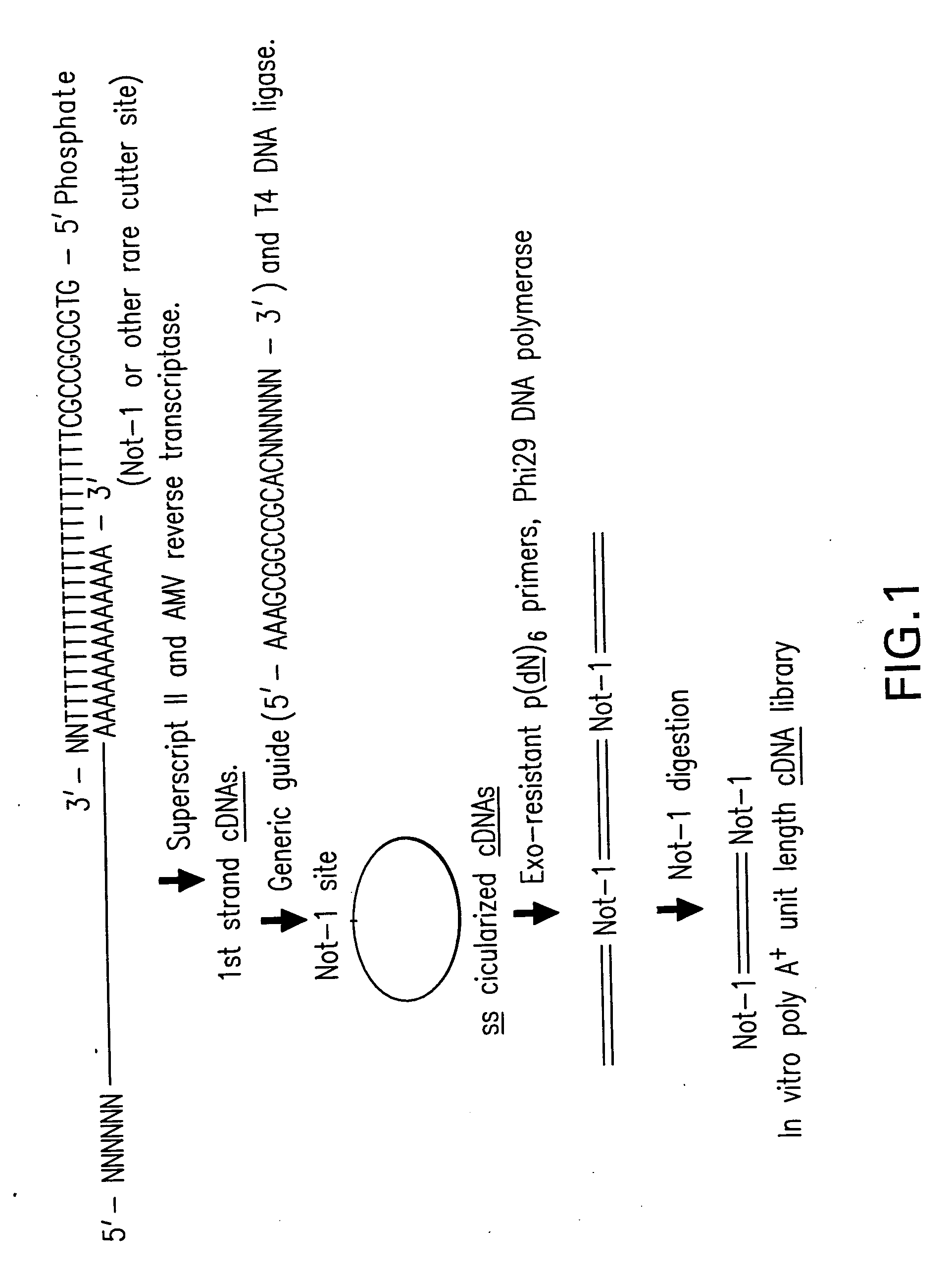
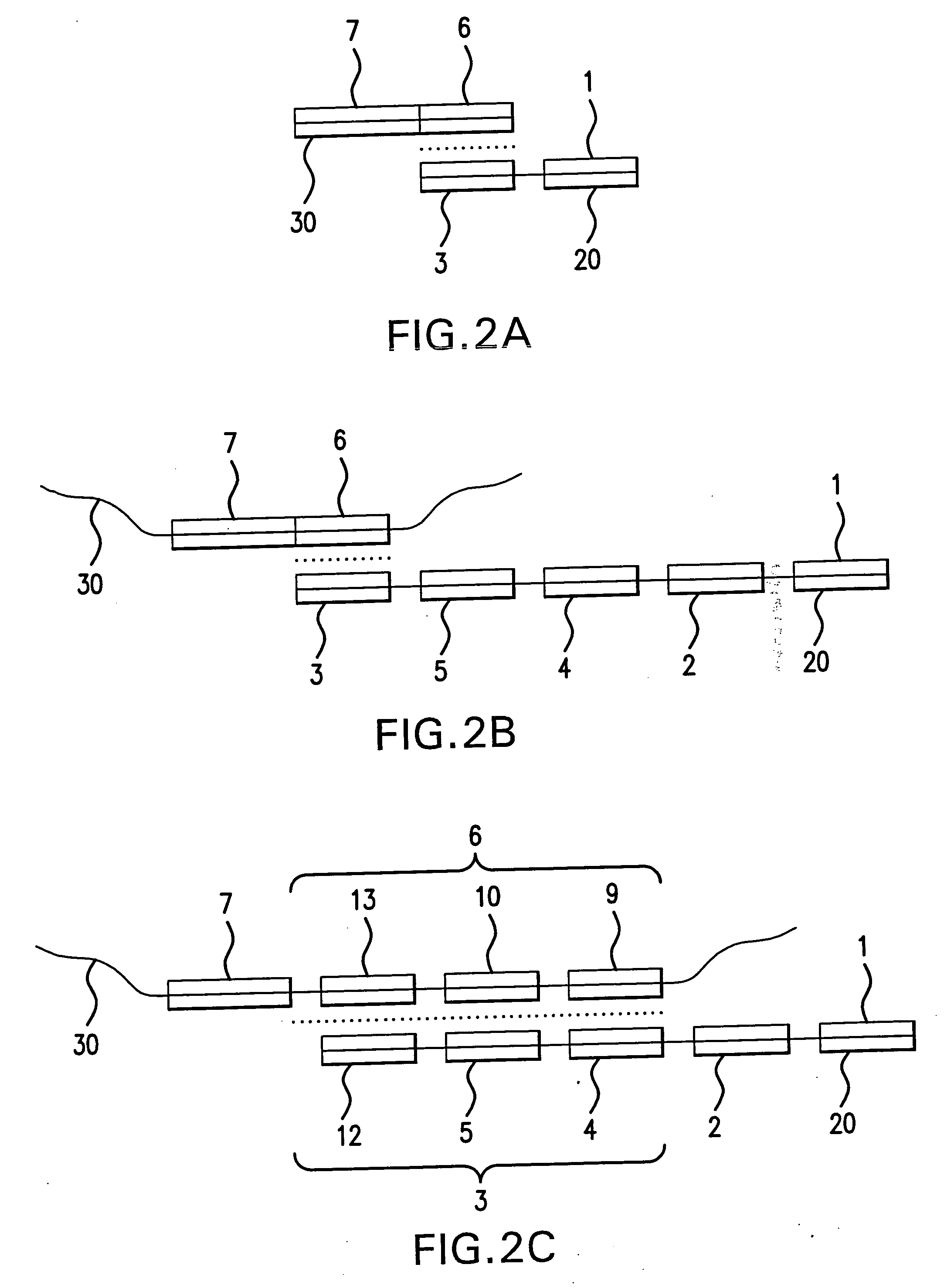
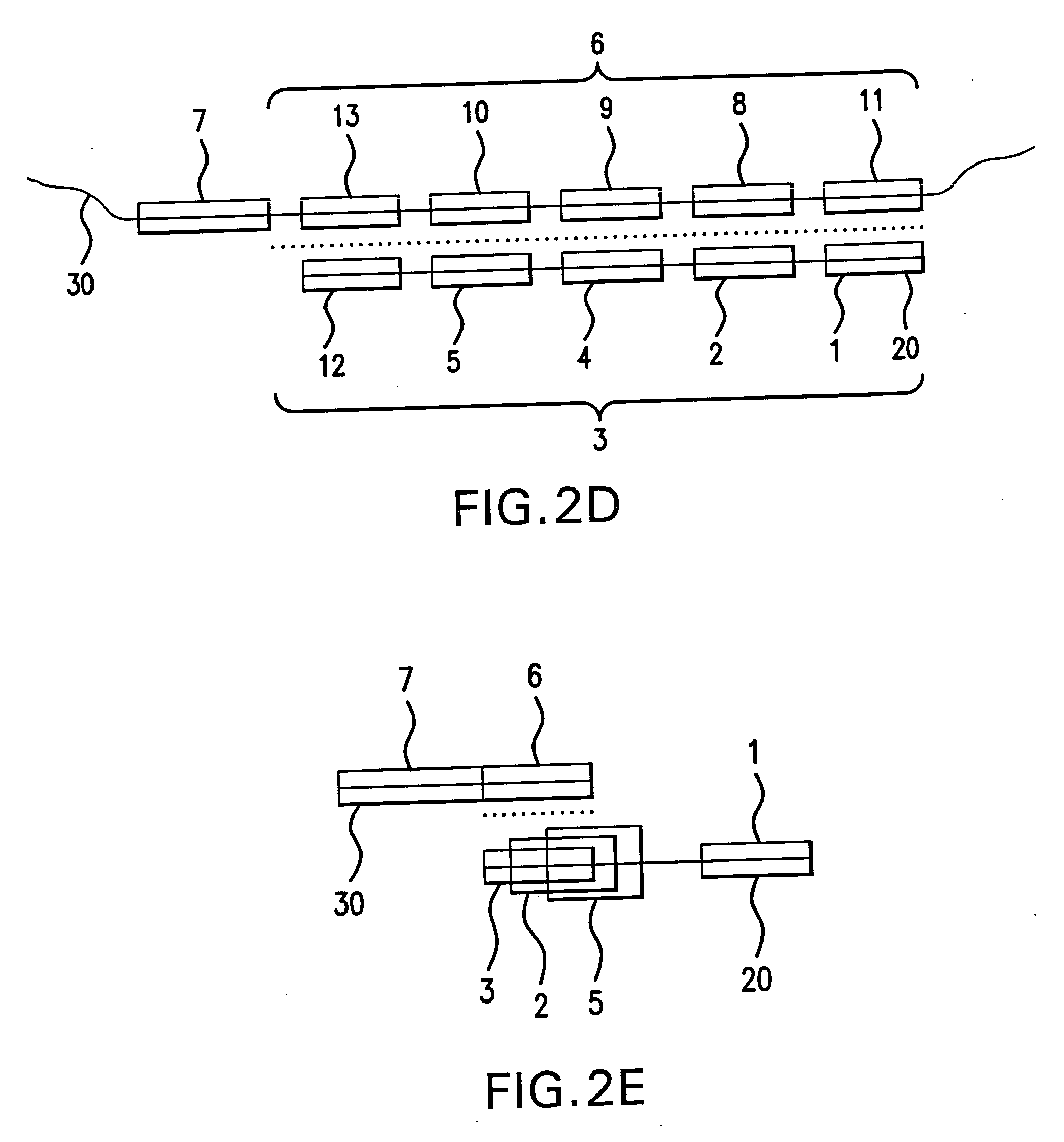
[0363] This example demonstrates a basic form of the disclosed method by amplifying polyadenylated RNA from a human adenorectal carcinoma. Control reactions indicate that amplification was dependent on circularization of first strand cDNA molecules.
[0364] First strand cDNA was synthesized from 1 μg poly A+ mRNA (human adenorectal carcinoma, SV-480) using a 5′-P-Not-1-(dT)20 cDNA primer (SEQ ID NO:1) and Superscript II reverse transcriptase. RNAse H+ reverse transcriptase was added to digest RNA templates. 0.1 M NaOH was then added and the mixture was incubated for 10 minutes. The reaction was then neutralized by adding 0.1 M HCl. The first strand cDNA molecules were purified using a Qiagen column to remove dNTPs and the primer. The first strand cDNA molecules were then circularized using T4 DNA ligase and a generic circularization probe (5′-AAAAGCGGCCGCACNNNNNN-3′; SEQ ID NO:2). The NNN segment of this circularization...
example 2
B. Example 2
Analysis of Amplification Products
[0366] Following the design concept (FIG. 1), first strand cDNA molecules were synthesized from 1 μg poly A+ mRNA from Adenocarcinoma cells (Clontech) using a 5′-phosphorylated cDNA primer as shown in FIG. 1 (oligo˜dT at the 3′-end and Not I endonuclease sequence on its 5′-phosphorylated end) using Superscript II reverse transcriptase (Invitrogen) at 42° C. for 1 hr followed by AMV reverse transcriptase (Invitrogen) at 37° C. for 1 hr. The first strand cDNA was isolated from the reaction mixture using Qiagen PCR product purification kit. First strand cDNA representing 100 ng mRNA ( 1 / 10th reaction product) was annealed to an excess of bridge oligonucleotide (circularization probe) (1 μg) in 20 μl×T4 DNA ligase buffer (heating to 80° C. followed by slow cooling to 30° C.) followed by the addition of 40U T4 DNA ligase for 12 hrs at 30° C. A fraction of the ligated cDNA reaction mixture, representing 5 ng mRNA, was then used for 100 μl amp...
example 3
C. Example 3
Analysis of Amplification Bias of Amplification Products
[0369] In order to determine the bias generated by amplification of a specific transcript from two different RNA samples, first strand cDNA synthesis was carried out from poly-A+ mRNA derived from human adult brain and fetal brain (Clontech). The first strand cDNA was ligated at 4 ng / μl concentration in the presence of a circularization probe. 10 ng of ligated first strand cDNA molecules were used for transcript amplification in a 30 μl reaction volume using Φ29 DNA polymerse and exonuclease-resistant p(dN)6 primers. To determine the relative abundance of DNA damage-inducible transcript (Dle3) targets per ng of the sample, Taqman assays were carried out using amplified and unamplified cDNA molecules representing adult and fetal brain mRNA. Transcript abundance in these samples was calculated by utilizing Taqman assay standard curve using varying amounts of genomic DNA (333 copies per ng genomic DNA). This procedure...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting temperature | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com