DNA sequencing method
a polynucleotide sequence and sequencing method technology, applied in the field of polynucleotide sequence determination, can solve the problems of high cost, slow, labor-intensive and expensive, and the technology used in the sequencing process, and achieve the effect of reducing the amount of subcloning and the number of overlapping sequences required
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example
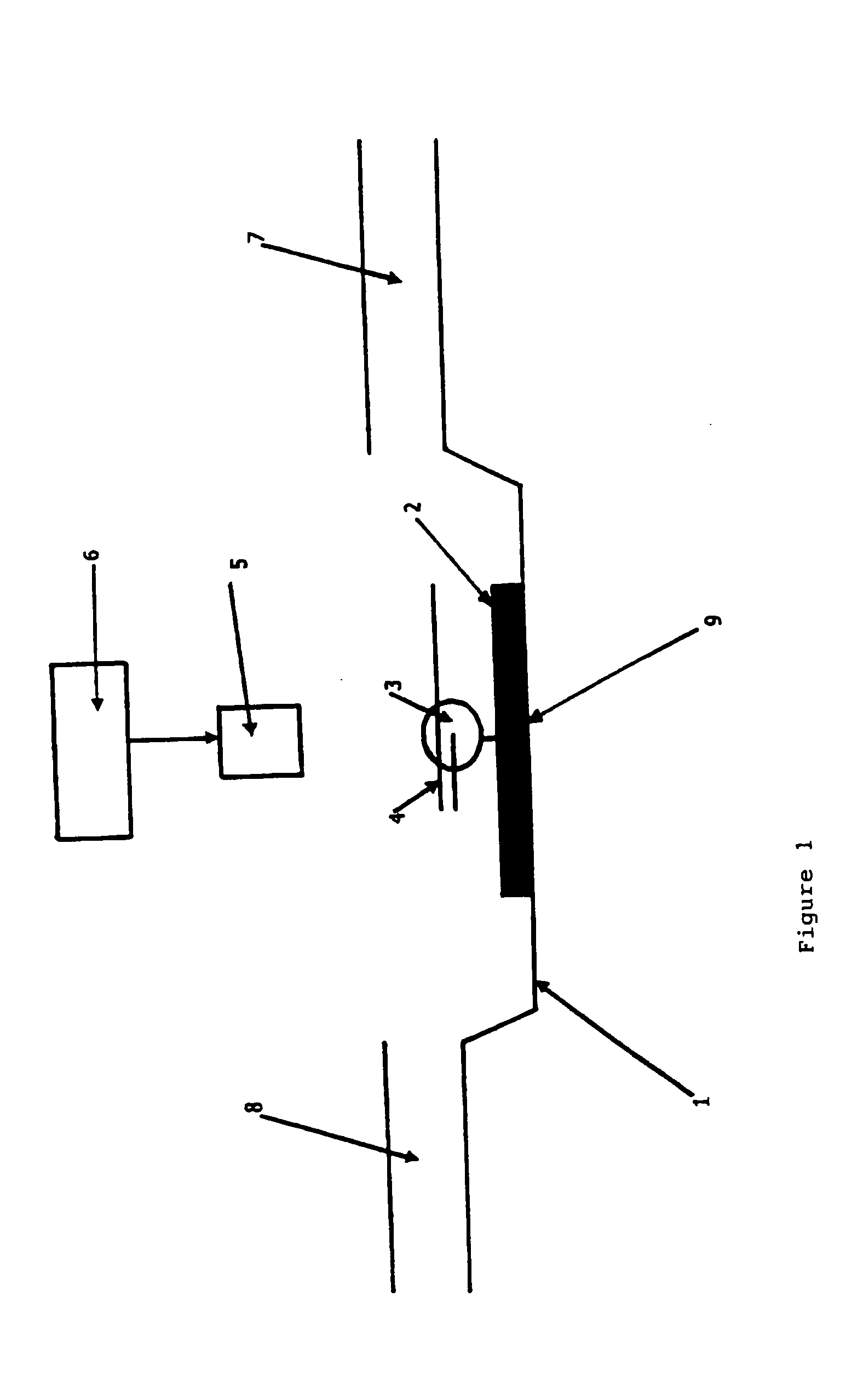
[0040] This Example used a confocal fluorescence setup, as shown in FIG. 1.
[0041] With reference to FIG. 1, the setup consists of a scan table (1) able to scan at high resolution in X, Y and Z dimensions, a class coverslip (2) which is part of a microfluidic flow cell system with an inlet (8) for introducing the primer-template polynucleotide complex (4) and nucleotides over the immobilised (9) polymerase molecule (3) within a buffer, and an outlet (7) for waste. Incident light from a laser light source (6) for donor excitation is delivered via an oil-immersion objective (5).
Protein Conjugation
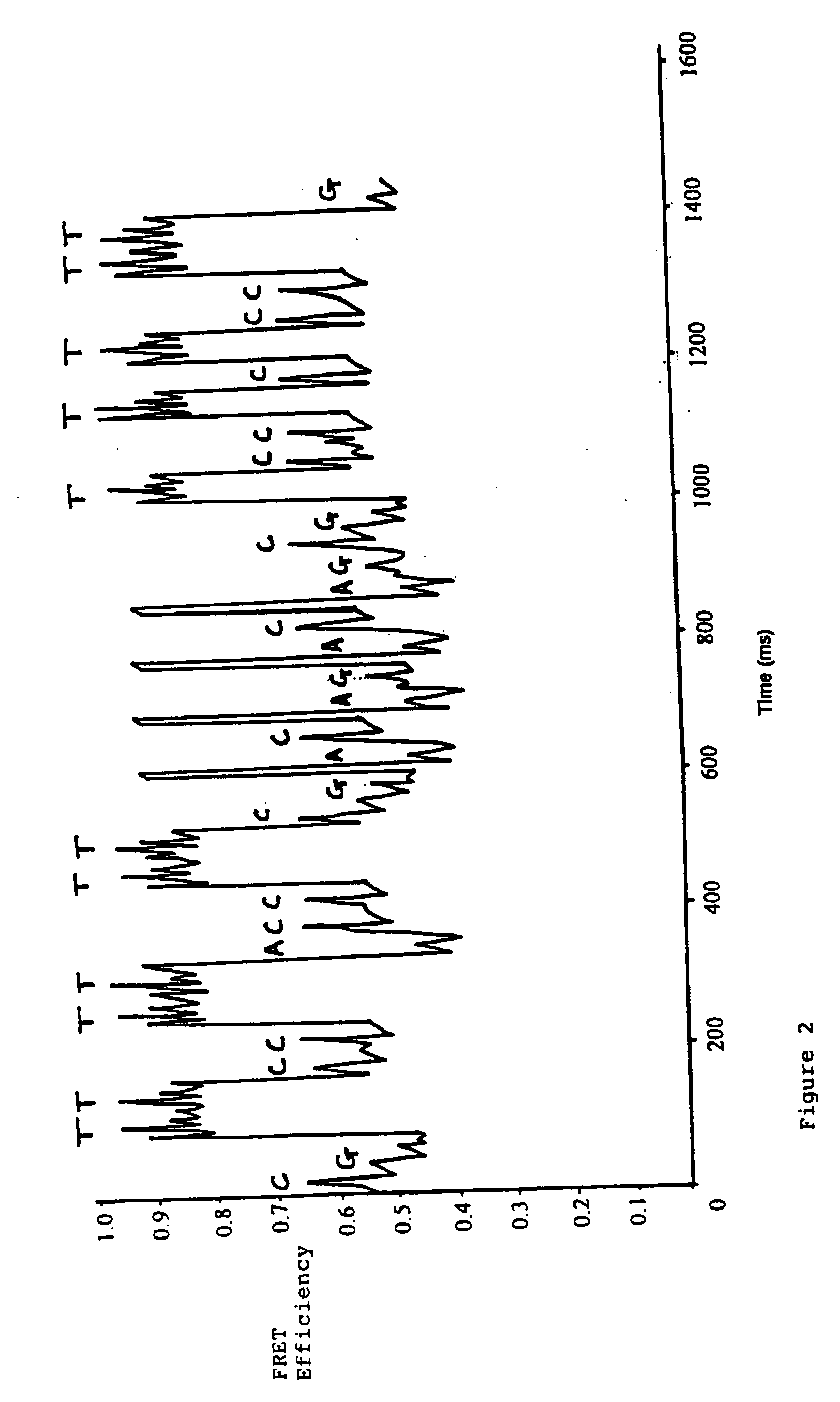
[0042] In this experiment, Tetramethylrhodamine (TMR, donor) and Cy5 (acceptor) where used as the FRET pair. This was due to their well separated emission wavelengths (>100 nm) and large Föster radius.
[0043] T7 DNA Polymerase from New England Biolabs (supplied at 10 000 U / ml) was used. 50 μl of T7 was buffer-exchanged in a Vivaspin 500 (Vivaspin) against 4×500 μl of 200 mM Sodium Acetate ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| emission wavelengths | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com