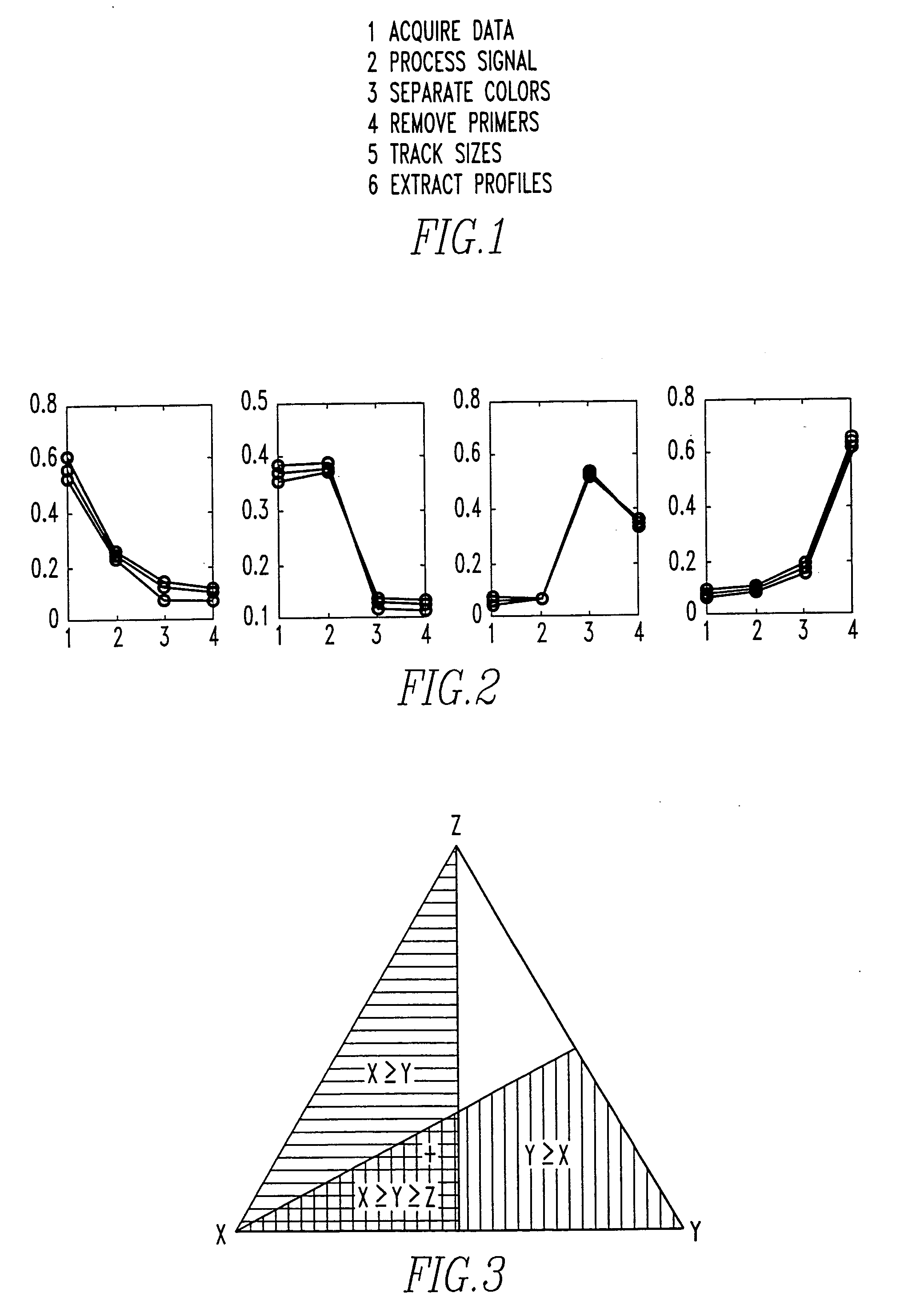
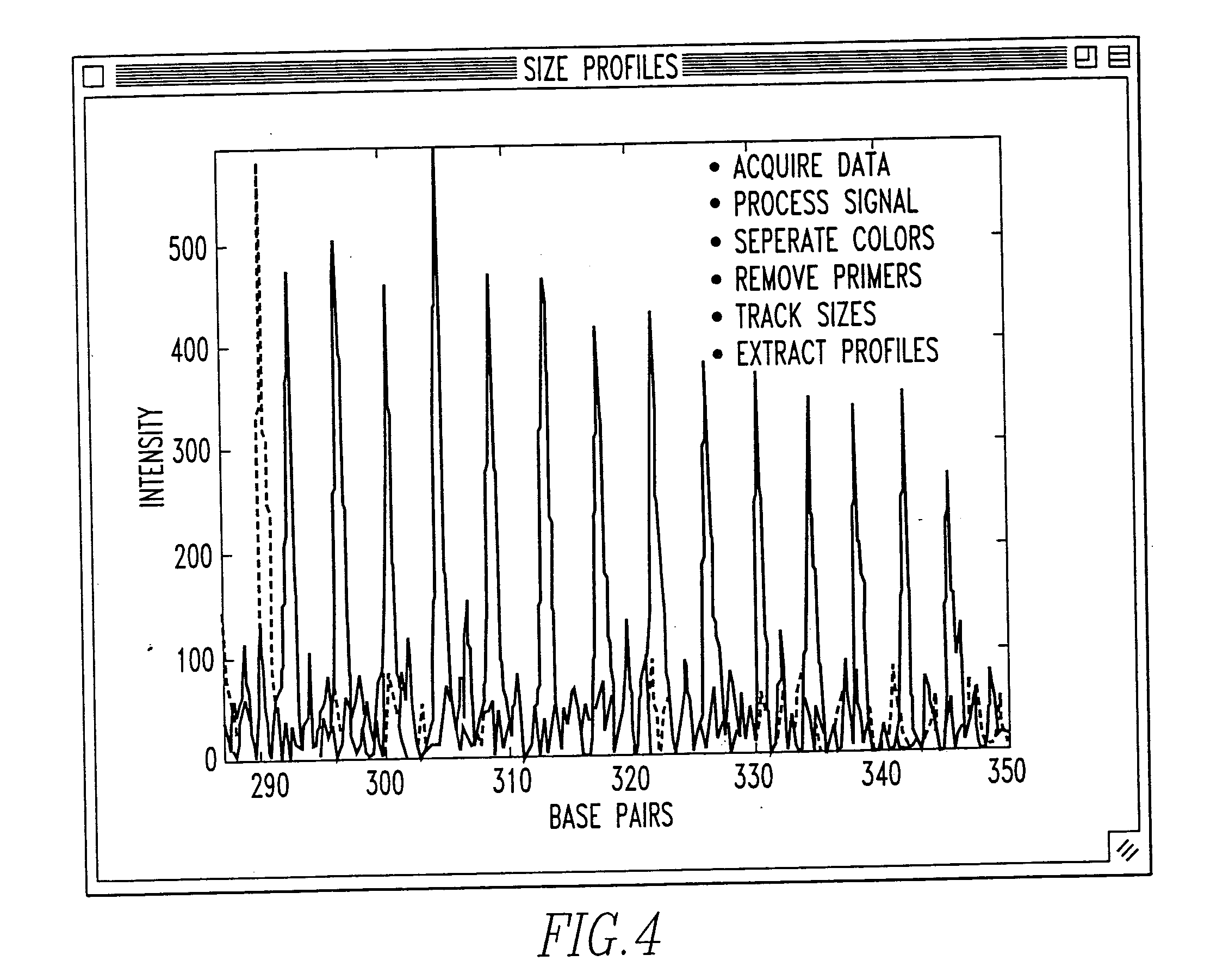
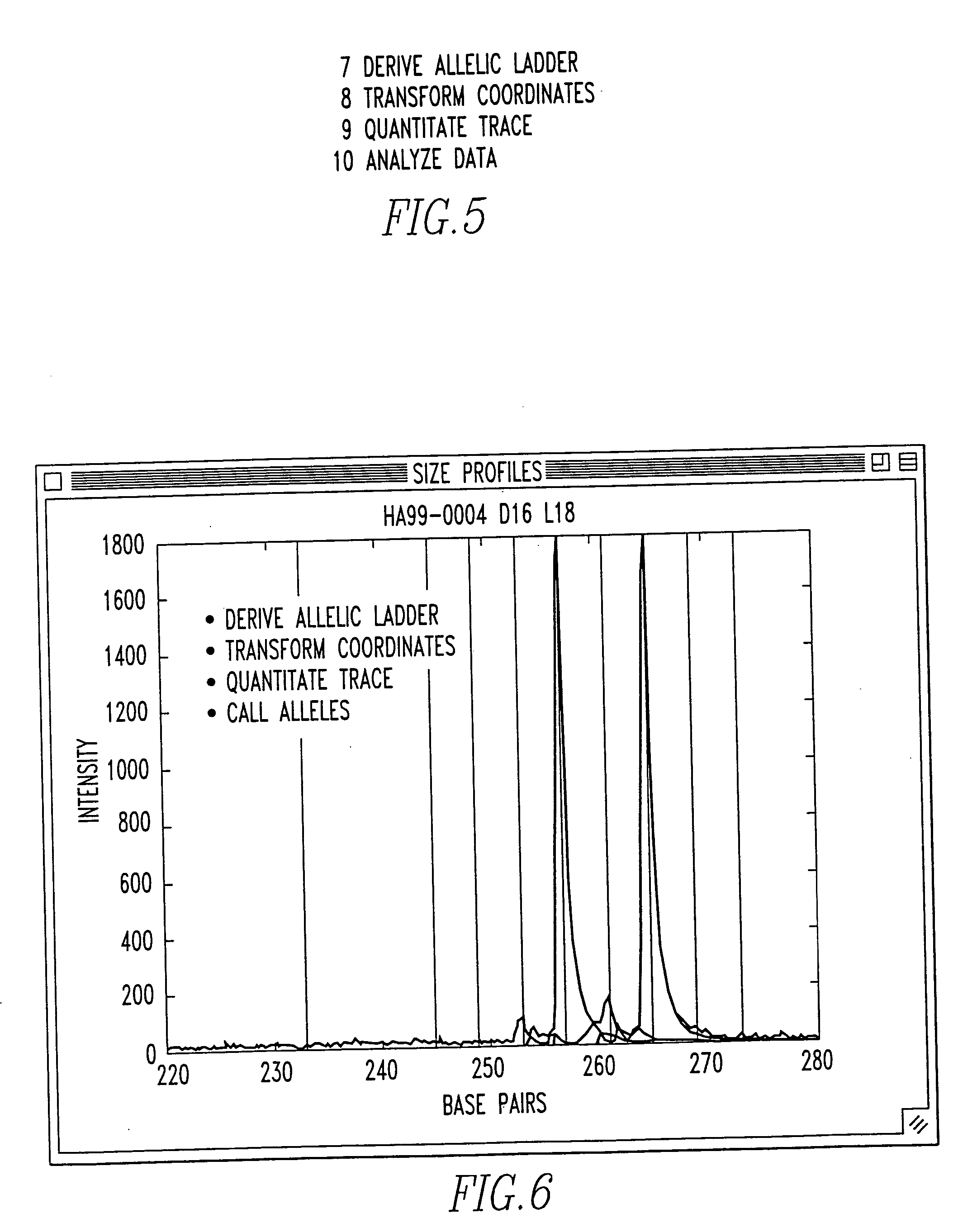
Method and system for DNA analysis
a dna analysis and method technology, applied in the field of dna analysis process, can solve the problems of over-examination of assay results, and inability to detect dna fragments in time,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
Data Generation
In the most preferred embodiment, genotyping data is generated using STR markers. These tandem repeats include mono-, di-, tri-, tetra-, penta-, hexa-, hepta-, octa-, nona-, deca- (and so on) nucleotide repeat elements. STRs are highly abundant and informative marker distributed thoughout the genomes of many species (including human). Typically, STRs are labeled, PCR amplified, and then detected (for size and quantity) on an electrophoretic gel.
The laboratory processing starts with the acquisition of a sample, and the extraction of its DNA. The extraction and purification are typically followed by PCR amplification. Labelling is generally done using a 5′ labeled PCR primer, or with incorporation labeling in the PCR. Prior to loading, multiple marker PCR products in k-1 different fluorescent colors are pooled, and size standards (preferably in a kth different color) is added. Size separation and detection is preferably done using automated fluorescent DNA sequence...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentrations | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com