Methods and systems for volume variation modeling in digital PCR
a volume variation and volume technology, applied in the field of methods and systems for volume variation modeling in digital pcr, can solve problems such as not yielding the correct number of targets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0014]To provide a more thorough understanding of various embodiments, the following description sets forth numerous specific details, such as specific configurations, parameters, examples, and the like. It should be recognized, however, that such description is not intended to limit the embodiments described to specific implementations, configurations, etc. Nor do the descriptions necessarily provide complete descriptions of the embodiments. As such, certain aspects, features, components, etc., may be omitted from the description of the various embodiments for ease of explanation.
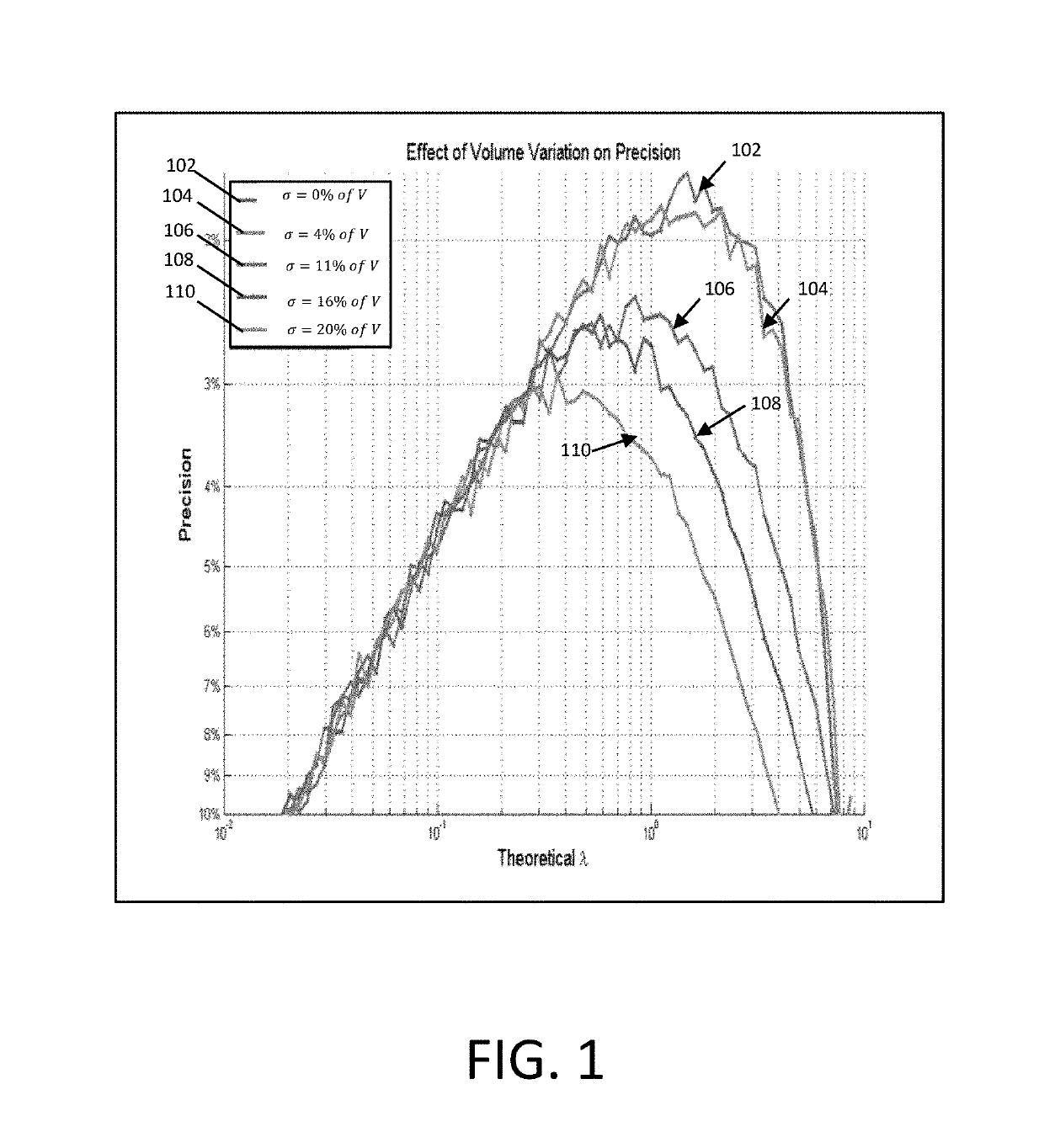
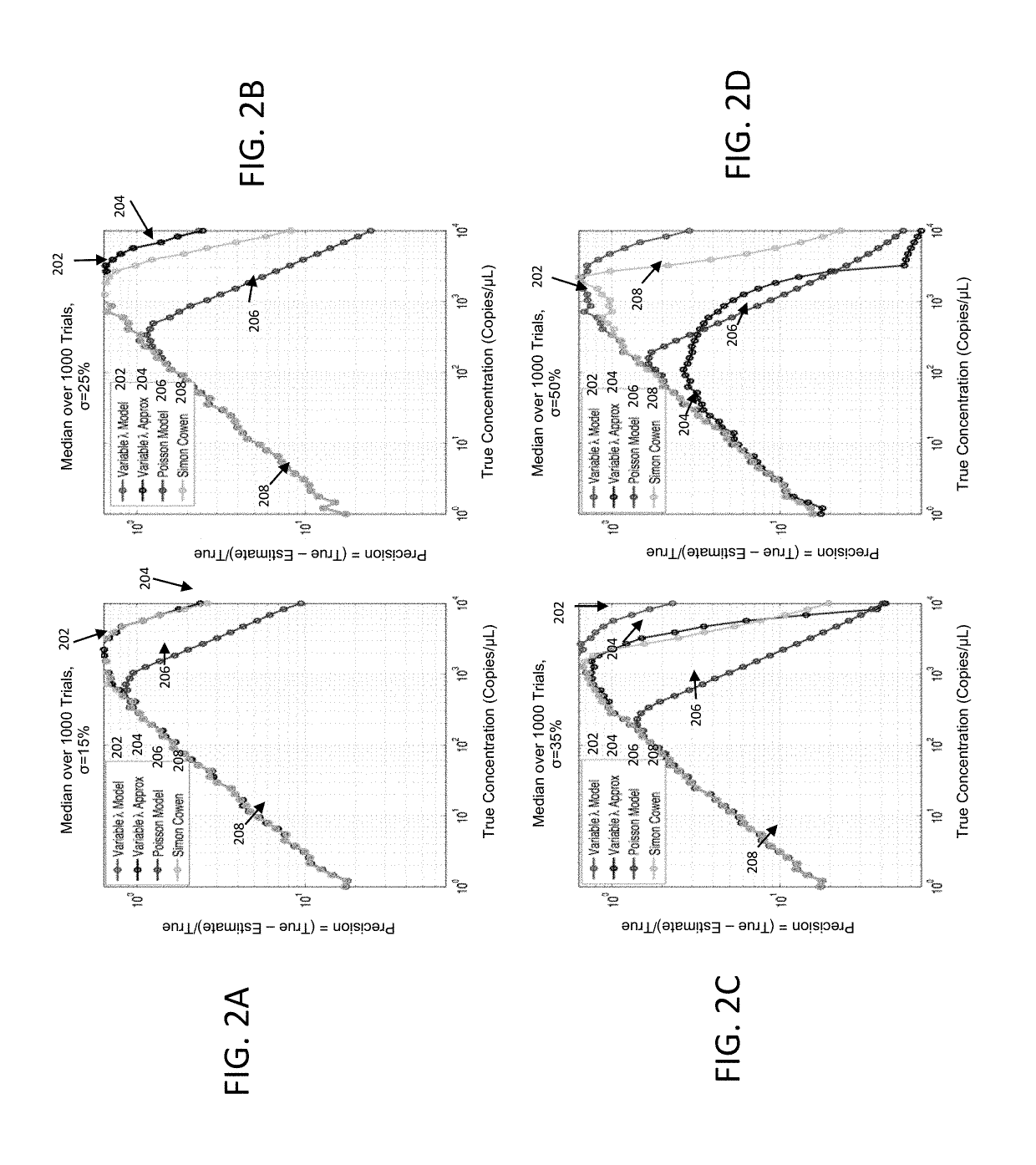
[0015]According to various embodiments described herein, a new quantification model that can be used to accommodate for volumetric variation and recover the quantification result at high precision is provided. Monte Carlo simulations are used to demonstrate the efficacy of the proposed model.
INTRODUCTION
[0016]In general, a digital PCR method distributes target molecules into a large number of partitions su...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| volumes | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com