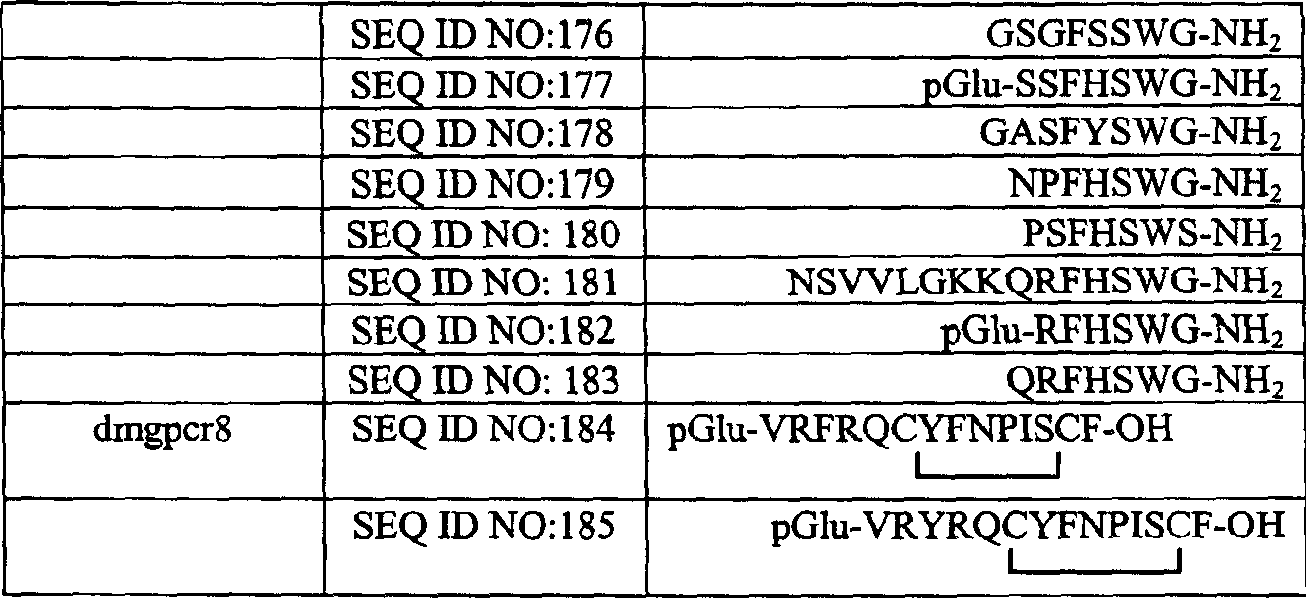
Drosophila g protein coupled receptors, nucleic acids, and methods related to the same
A Drosophila and lateral body technology, applied in Drosophila G protein-coupled receptors, nucleic acid and related fields, can solve the problem of unidentified insect C-type pharyngeal body inhibin
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0289] Example 1: Identification of DmGPCR
[0290] Use various gene search software tools to convert the Celera genotype Drosophila database into a predicted protein and mRNA database to predict the mRNA to be produced ("PnuFlyPep" database). Methods for analyzing genome databases by gene search software tools are known to those skilled in the art.
[0291] The nucleotide sequences of several C. elegans FaRP GPCRs were used as query sequences for the above-mentioned mRNA database. Various tools, including FASTA and Gap BLAST, were used to search for similar regions in the database (Altschul et al., Nuc. Acids Res., 1997, 25, 3389, which is fully incorporated herein for reference).
[0292] To put it simply, the BLAST algorithm, which represents a basic local arrangement comparison search tool, is suitable for determining sequence similarity (Altschul et al., Journal of Molecular Biology 215: 403-410 (1990), which is fully incorporated herein for reference). National Center for Bi...
Embodiment 2
[0295] Example 2: Cloning of DmGPCR
[0296] cDNA preparation
[0297] From adult fruit fly (Drosophila melanogaster) poly A + CDNA was prepared from RNA (Clontech Laboratories, PaloAlto, CA) or adult Drosophila total RNA (below). To obtain total RNA, freeze anesthetize the parent stock of Drosophila (Biological Supply Company, Burlington, NC), which contains 10ml H 2 O, 10ml Formula4-24 Instant Drosophila medium and 6-10 active dry yeast (Biological Supply Company) culture tube add 5-6 adult fruit flies. A polyurethane foam plug is placed at the end of each test tube, and the fruit flies are incubated at room temperature for 4-6 weeks. At the maturity stage, the test tube is frozen, and the anesthetized fruit flies are poured into a 50 ml polypropylene test tube in liquid nitrogen. Frozen fruit flies are stored at -70°C until they are ground in a mortar and mashed with a pestle in the presence of liquid nitrogen. Decant the powdered tissue and some liquid nitrogen into a 50ml pol...
Embodiment 3
[0336] Example 3: Northern blot analysis
[0337] Northern blot analysis can be performed to detect mRNA expression. As described above, using sense-oriented oligonucleotides and antisense-oriented oligonucleotides as primers, a part of SEQ ID NO: 1, 3, 5, 7, 9, 11, 13, 15, was amplified, The GPCR cDNA sequence of the nucleotide sequence of 17, 19, 21, and 23.
[0338] Several human tissue Northern blots from Clontech (human II#7767-1) were hybridized with the probe. The pre-hybridization was performed at 42°C, 5xSSC, 1x Denhardt's reagent, 0.1% SDS, 50% formamide, and 250 mg / ml salmon sperm DNA for 4 hours. Hybridization was carried out overnight at 42°C in the same mixture, adding about 1.5×10 6 cpm / ml labeled probe.
[0339] The probe was labeled with α-32P-dCTP by Rediprime DNA labeling system (Amersham Pharmacia), purified on a Nick column (Amersham Pharmacia), and added to the hybridization solution. Wash the filter several times in 0.2xSSC and 0.1% SDS at 42°C. At -80°C, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com