Molecular method for identifying Mucomyzius japonicus in North China of China
A kind of hibiscus, auxiliary identification technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
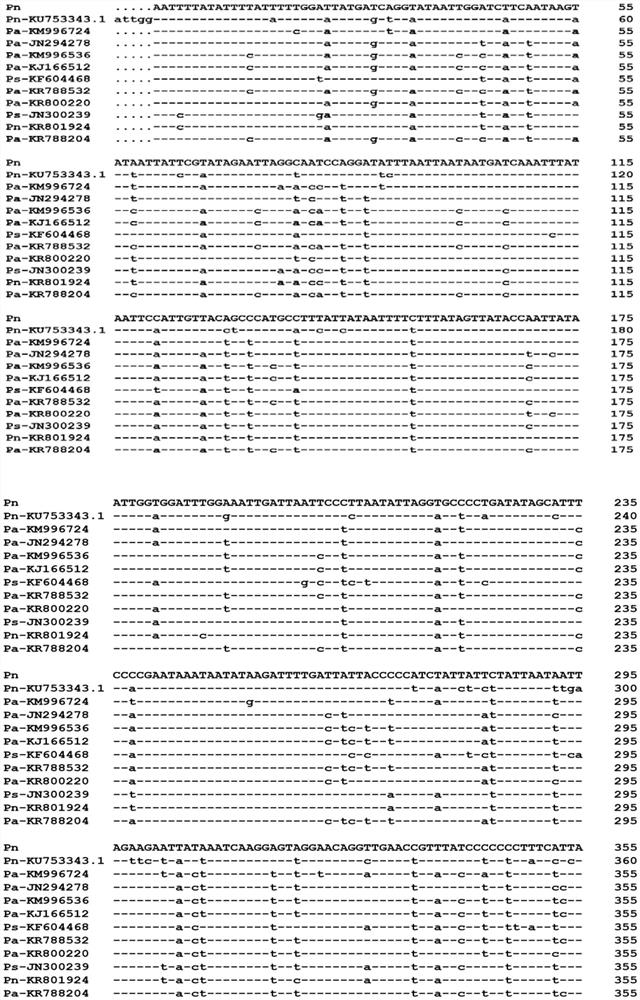
[0045] Example 1. Obtaining of the mitochondrial COI gene of A. japonica
[0046] 1. DNA extraction
[0047] Extraction and selection of well-preserved 2 samples of A. japonica from Luquan District, Shijiazhuang, North China, and 1 sample of . Perform single-head DNA extraction. The concentration and quality of genomic DNA were detected by NanoDrop Spectrophotometer (DS-11Envivx), the integrity of genomic DNA was detected by 1% agarose gel electrophoresis, and stored at -20°C for future use.
[0048] 2. Amplification of mitochondrial COI gene fragments
[0049] Using the genomic DNA in 1 above as a template, perform PCR amplification with the following general primers to obtain PCR amplification products.
[0050] The PCR amplification of the above-mentioned mitochondrial COI gene fragment adopts universal primers,
[0051] Upstream primer LCO1490: GGTCAACAAATCATAAAGATATTGG;
[0052] Downstream primer HCO2198: TAAACTTCAGGGTGACCAAAAATCA.
[0053] The PCR amplification sys...
Embodiment 2
[0069] Example 2, Application of Mitochondrial COI Gene in Identifying A. japonica
[0070] 1. Genomic DNA extraction
[0071] Extract 3 Pimpla nipponica, 3 Pimpla disparis, and 3 Pimpla disparis from Luquan District of Shijiazhuang and Fudong District of Handan City (2018 and 2019) Genomic DNA of a bee (Diplazonlatatorius).
[0072] 2. Amplification of mitochondrial COI gene
[0073] Using the above-mentioned genomic DNA as a template, the following general primers were used to amplify according to the method of Example 1:
[0074] Upstream primer LCO1490: GGTCAACAAATCATAAAGATATTGG;
[0075] Downstream primer HCO2198: TAAACTTCAGGGTGACCAAAAATCA.
[0076] Obtain the PCR amplification product.
[0077] PCR amplification products of different bees were sequenced.
[0078] The result is as follows:
[0079] The sequencing results of the three A. japonicus were sequence 1;
[0080] The sequencing results of the three gypsy moths were all sequence 3;
[0081] The sequencing...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com