TE molecular marker closely linked with peach double petals and application thereof
A technology of molecular markers and double petals, which is applied in the direction of recombinant DNA technology, microbiological determination/inspection, DNA/RNA fragments, etc., can solve the problems such as the difficult molecular identification of peach double petals, achieve good application and promotion prospects, and have few restrictions , the effect of the simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. Development of double flower gene markers in natural variety populations
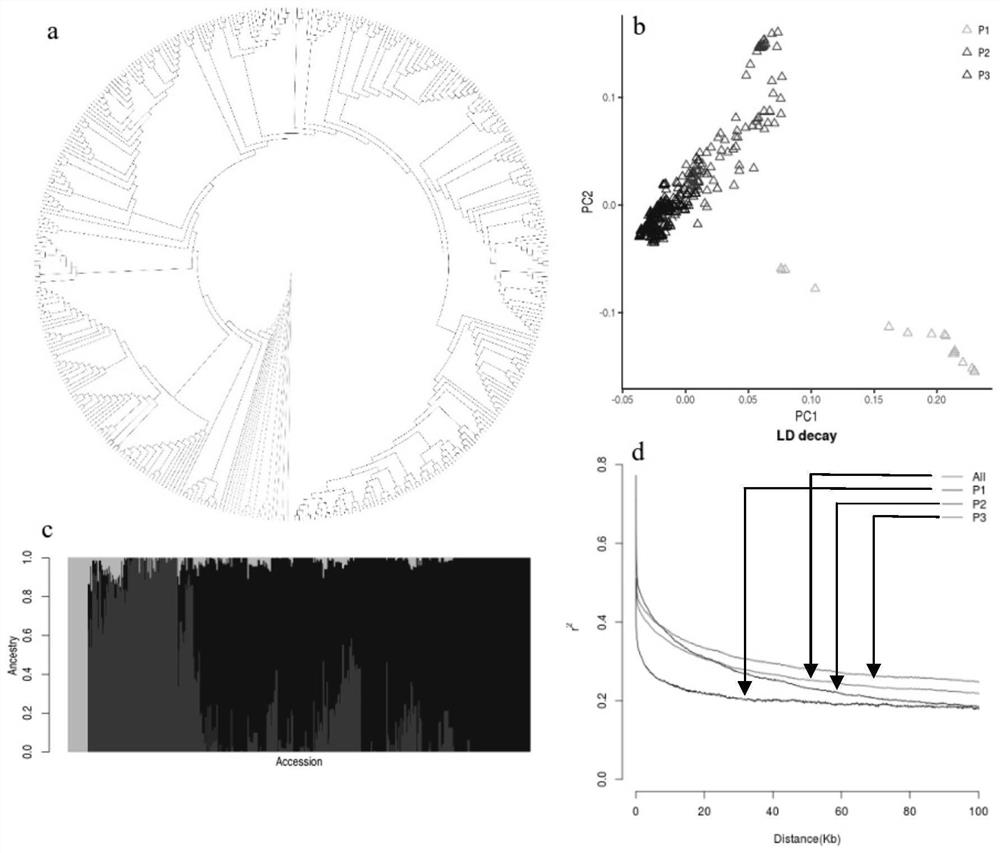
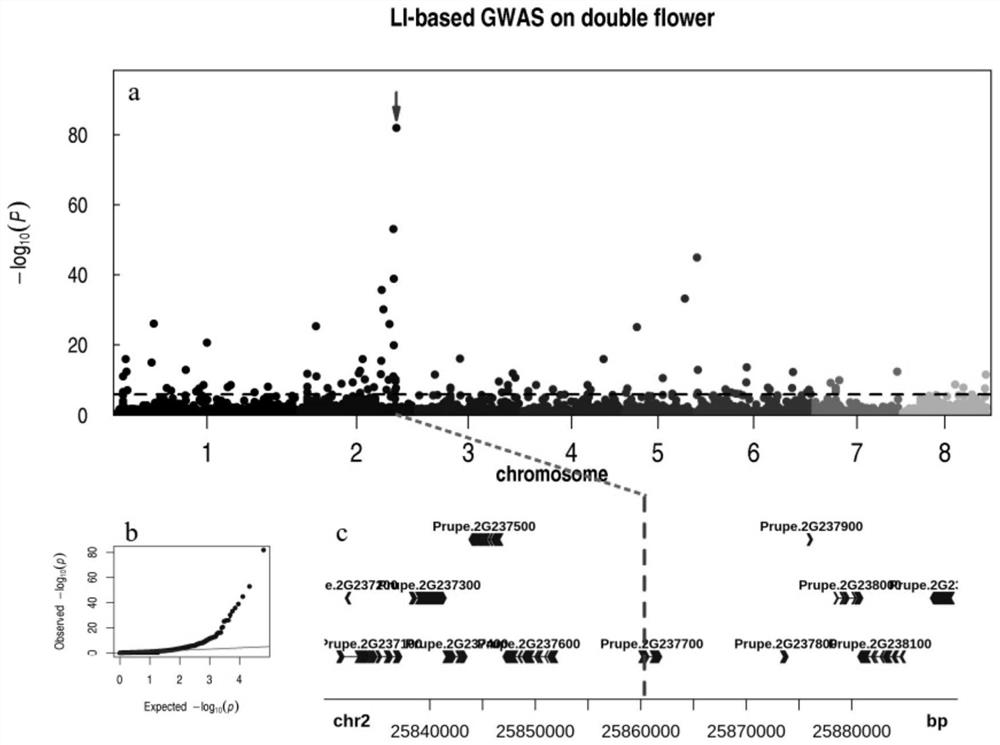
[0029] In order to solve the problem of obtaining accurate phenotype-associated sites, the present invention combined the high-depth resequencing data (average depth 20.3X) of 417 cultivated peach varieties covering each major production area for structural variation (SVs) detection, and screened out SVs sites~ 64,000. Using genome-wide association analysis (GWAS), it was found that a TE located at 25,860,343bp of chromosome Pp02 in the peach genome was strongly related to double flowers, named FD-TE-25.86. Figure 4 at the box.
[0030] figure 1 The phylogenetic tree, PCA (principal component analysis), population structure and LD (linkage disequilibrium) reduction trend of 417 peach resources are shown in figure 1 The a in is the high-quality SNP variation data filtered based on the whole genome, and the phylogenetic tree constructed based on the proximity method. figure 1 b. is the PCA p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com