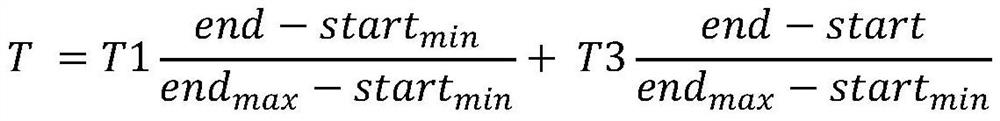A method for decompressing DNA self-indexed intervals
A decompression and self-indexing technology, applied in the direction of code conversion, electrical components, etc., can solve the problems of long decompression time and large data storage space, and achieve the effect of reducing decompression time, strong applicability and storage space
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
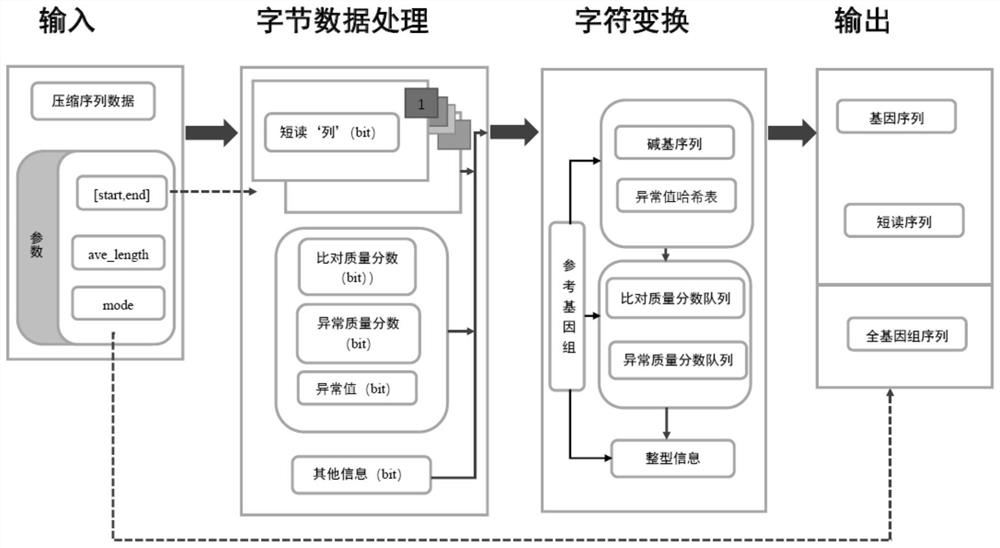
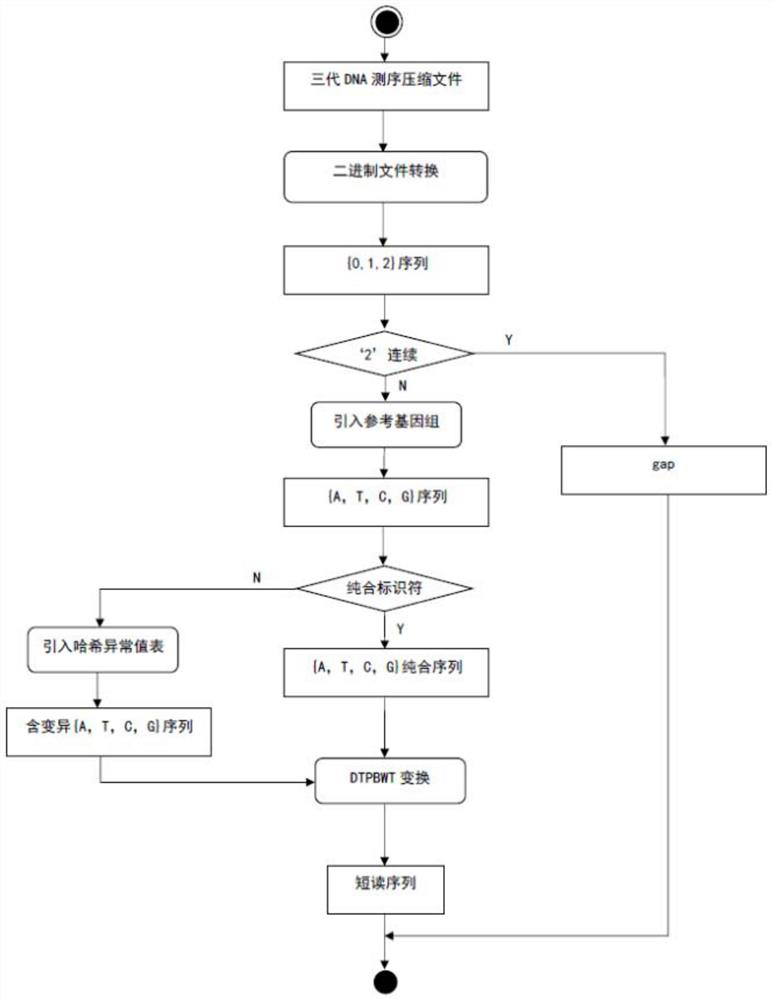
[0015] Specific implementation mode one: combine figure 1 This embodiment will be described. A DNA self-index interval decompression method described in this embodiment, the method is specifically implemented through the following steps:
[0016] Step 1. Input the sequence data file to be decompressed, and configure the index interval parameter ([start, end]) and the decompression output mode parameter (mode);
[0017] Step 2, according to the index interval parameter, determine the interval range that needs to be decompressed in the sequence data file to be decompressed;
[0018] Step 3. According to the header file information of the sequence data file to be decompressed, determine the sequenced short-read base bit information (short-read "column" for short) within the range that needs to be decompressed, which can be compared to the bases on the reference genome. Sequencing quality score bit information (referred to as comparison quality score), sequencing quality score b...
specific Embodiment approach 2
[0030] Embodiment 2: This embodiment is a further detailed description of Embodiment 1. The decompression output mode parameter determines the type of data to be decompressed and output.
specific Embodiment approach 3
[0031] Specific embodiment three: this embodiment is a further specific description of specific embodiment two. When the decompression output mode parameter is set to 1, the data type of the decompression output is a gene sequence. When the decompression output mode parameter is set to When 2, the decompressed output data type is a short-read sequence. When the decompressed output mode parameter is set to 3, the decompressed output data type is a whole genome sequence.
[0032] By default it is 0, which decompresses according to the short-read sequence condition.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com