Molecular marker for detecting deep notch marker character of rape leaf margin and application thereof
A technology related to molecular markers and rapeseed leaves, which is applied in the direction of microbial measurement/inspection, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of successful gene transfer of the leaf edge deep notch trait of rapeseed, and achieve remarkable results. High light efficiency, remarkable resistance to dense planting, and the effect of overcoming low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] 1. Construction of rapeseed with leaf margin deep notch marker traits and location of rapeseed leaf margin deep notch marker traits
[0036] Brassica oleracea Zhongshuang 11 was crossed with a wild plant, Rapeseed brassicae, and a rapeseed with deep notched leaf margins was obtained. The leaves with deep notches in rapeseed leaf margins were analyzed by full-length transcriptome sequencing (iso-seq), and the genes that were transferred from the saponinia genome to the rapeseed genome that caused the deep-cleft phenotype of rapeseed leaves were excavated. The specific process is:
[0037] A total of 92,897 full-length transcripts were detected in rapeseed with deep notch marks on leaf margins. Using the genome sequenced on Shuang11 in the parents as the reference genome, a total of 80,879 transcripts were compared to the reference genome. It was 87.06%, and the remaining 12018 transcripts were not aligned to the reference genome. The genomes of cabbage and cabbage were...
Embodiment 2
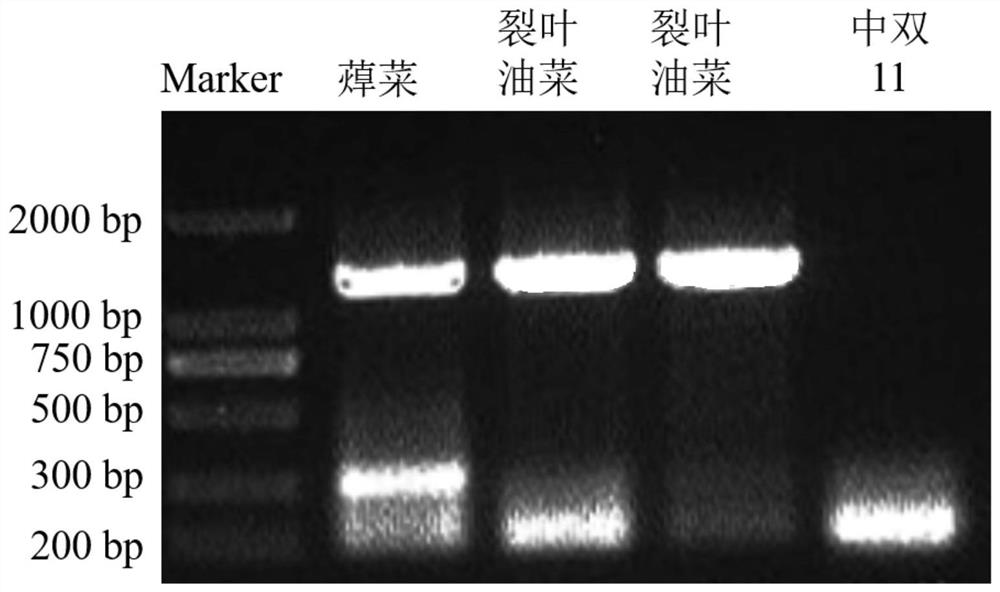
[0059] Example 2 Comparative Example of Molecular Markers of Leaf Margin Deep Notches Marking Traits
[0060] Using exactly the same steps as in Example 1 to obtain rapeseed with deep notch markers on leaf margins, and locate the transcripts of deep notch markers on rapeseed leaf margins, the cDNAs of transcripts that are not similar to Chinese cabbage, cabbage and rapeseed sequences were screened The sequence is shown in SEQ ID NO: 6, the nucleotide sequence translated from the transcript is shown in SEQ ID NO: 7, and the amino acid sequence of the translated protein is shown in SEQ ID NO: 8.
[0061] According to the sequence of SEQ ID NO: 6, a primer pair of the sequence is designed and synthesized, and the primer pair is composed of single-stranded DNA molecules shown in SEQ ID NO: 9 and SEQ ID NO: 10.
[0062] The sequence shown in SEQ ID NO: 6 was named LSD1, and the sequence shown in SEQ ID NO: 5 was named LSD2. The proteins translated from the above sequences were comp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com