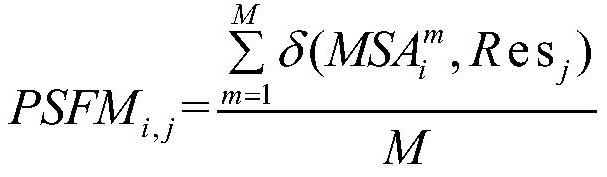Molecular virtual screening method based on protein sequence alignment
A protein sequence, virtual screening technology, applied in the fields of bioinformatics and computer applications, can solve the problem that the virtual screening method cannot work well
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0035] The present invention will be further described below in conjunction with the accompanying drawings.
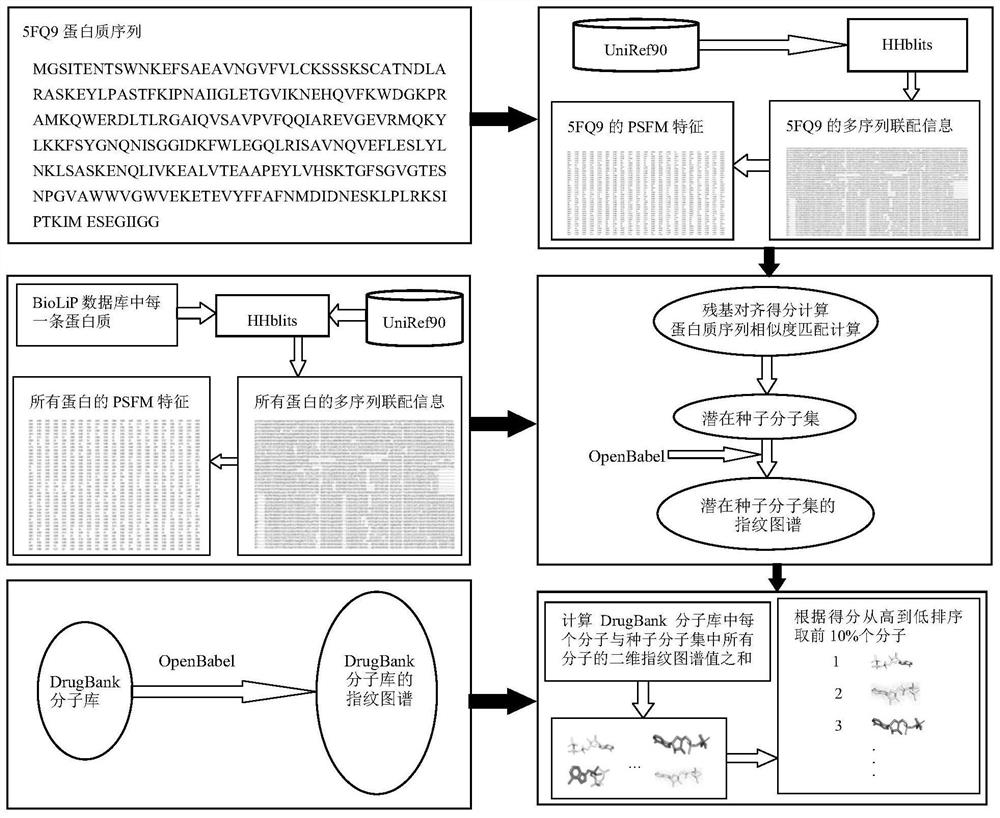
[0036] refer to figure 1 and figure 2 , a molecular virtual screening method based on protein sequence alignment, comprising the following steps:
[0037] 1) Input a protein sequence P to be subjected to molecular screening with the number of residues L;
[0038] 2) For the protein sequence P, use the HHblits (https: / / toolkit.tuebingen.mpg.de / # / hhblits) program to search the database UniRef90 (ftp: / / ftp.uniprot.org / pub / databases / uniprot / uniref / uniref90 / ), generating a multiple sequence alignment information comprising M sequences, denoted as MSA;
[0039] 3) For the MSA file, calculate the position-specific frequency matrix whose size is L×20, denoted as PSFM:
[0040]
[0041] Among them, PSFM i,j Represents the element in row i and column j in PSFM, i=1,2,...,L,j=1,2,...,20, Res j Indicates the jth of 20 residues (A, R, N, D, C, Q, E, G, H, I, L, K, M, F, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com