Application of Sir2 family gene or protein in regulation and control of sizes of plant organs
A plant organ, gene technology, used in applications, plant products, genetic engineering, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Cloning and sequence analysis of LcSRT1 and LcSRT2 genes
[0028] The present invention uses the previously obtained transcriptome data of Litsea cubeba (see accessionnumber SRA080286 in the SRA database of NCBI), and screens out two transcripts whose annotation information is Sir2 family protein, and both transcripts have completely open reads frame (ORF). The primers of two genes were designed according to the ORF sequence, and the Sir2 family genes were cloned from Litsea cubeba by RT-PCR. The Sir2 family genes are named as LcSRT1 and LcSRT2, which have 1,359bp (full CDS sequence as the nucleotide sequence shown in Seq ID No: 1) and 1,158bp (full CDS sequence as the nucleotide sequence shown in Seq ID No: 2) respectively. acid sequence), encoding 452 amino acids (amino acid sequence as shown in Seq ID No: 3) and 385 amino acids (amino acid sequence as shown in Seq ID No: 4), respectively.
[0029] Among them, the CDS amplification primer of LcSRT1 gene is...
Embodiment 2
[0035] Example 2 Overexpression of LcSRT1 and LcSRT2 genes can increase plant vegetative organs
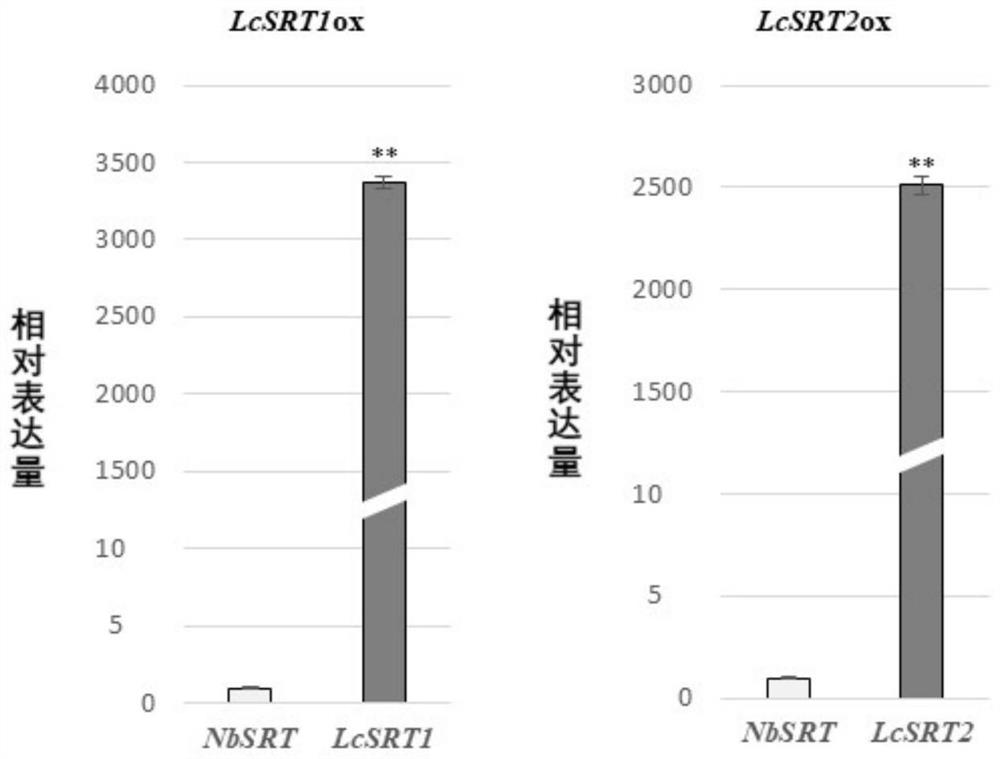
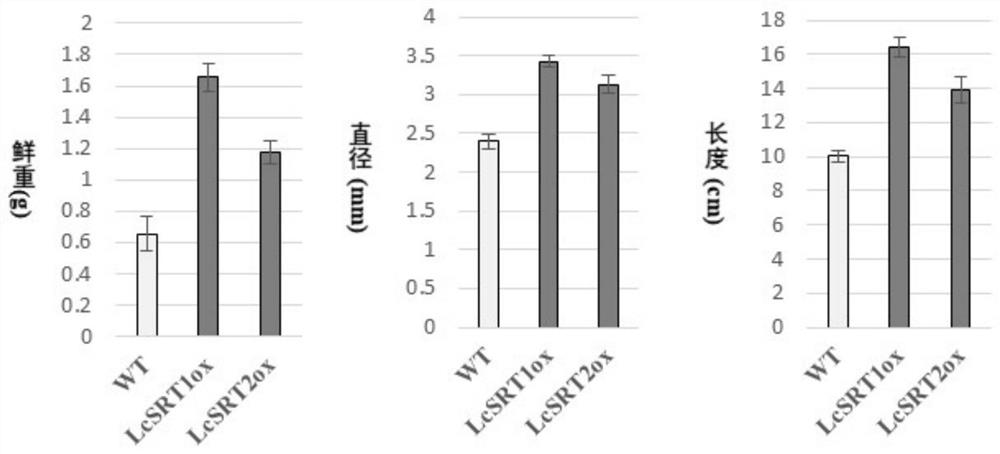
[0036] The LcSRT1 and LcSRT2 genes obtained in Example 1 were heterologously expressed in tobacco (N. benthamiana), and the present invention found that in the respective positive transgenic plants, the expression levels of the two genes increased by 3,373% compared with the SRT gene in tobacco. times and 2,512 times, such as figure 1 shown. At the same time, the present invention also found that the transgenic positive plants of the two genes had a phenotype of enlarged plant organs compared with the wild type, which was reflected in the enlarged leaves, increased stem diameter (also called increased stem diameter), and increased stem diameter at the same developmental stage. A phenotype of increased stem height (also called increased length) and increased fresh weight of the plant.
[0037] Specifically: heterologous expression of LcSRT1 and LcSRT2 genes in tobacco. After pos...
Embodiment 3
[0039] Example 3 The overexpression of LcSRT1 and LcSRT2 genes can increase the production of plant monoterpenes
[0040] After the LcSRT1 and LcSRT2 genes obtained in Example 1 were heterologously expressed in tobacco (N. benthamiana), the present invention found that in the respective positive transgenic plants, the expression levels of the two genes were respectively increased compared with the SRT gene in tobacco. 3,373 times and 2,512 times, such as figure 1 shown. The present invention uses the headspace-SPME-GC-MS method to comparatively analyze the chemical components of the essential oils in LcSRT1ox, LcSRT2ox and wild-type plant WT, and specifically finds that (with figure 2 shown):
[0041] Compared with control wild-type plants, the overexpression of LcSRT1 and LcSRT2 genes can significantly increase the production of (1R)-(+)-α-pinene, D-limonene, cineole, β-ring citral and geranial. Metabolites are important constituents of monoterpenes in essential oils.
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com