A kind of primer combination, probe combination and its application in detecting porcine virus, detection reagent, kit and detection method
A detection reagent and primer combination technology, applied in the field of probe combination and its application in the detection of porcine virus, and primer combination, can solve the problems of difficult clinical sample detection, and achieve improved detection efficiency, cost saving, and simple operation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1: This embodiment provides a method for joint detection of five porcine viruses. Specifically, a five-fold fluorescent quantitative PCR detection method was established to establish a joint detection method for five porcine viruses.
[0069] 1. Design of primers and TaqMan probes
[0070] The B646L gene sequences of different African swine fever virus (ASFV) strains published by GenBank (accession numbers: MN393476.1, MN715134.1, MN172368.1, LR743116.1, MK333180.1, MH910495.1, MN336500.2); 3D gene sequences of foot-and-mouth disease virus (FMDV) strains (accession numbers: FJ175661.1, GU384683.1, DQ989323.1, DQ989308.1, FJ175662.1, FJ175665.1, MF372126.1, DQ989320. 1); Nsp2 gene sequence of porcine reproductive and respiratory syndrome virus (PRRSV) strain (accession numbers: NC_043487.1, EF635006.1, EF112445.1, EF641008.1, AY150564.1, MH500776.1, MN119307.1 , KP771777.1); 5´UTR gene sequence of classical swine fever virus (CSFV) strain (accession numbers:...
Embodiment 2
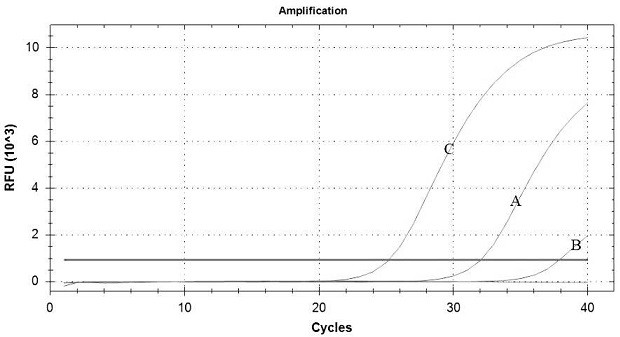
[0127] Example 2: Sensitivity Verification of Five-fold Fluorescent Quantitative PCR Detection Method
[0128] ASFV (5.8 × 10 4 copies / μL), FMDV (8.1×10 4 copies / μL), CSFV (5.1×10 4 copies / μL), HP-PRRSV (9.7×10 4 copies / μL), PRV wild virus (7.6×10 4 copies / μL) Take 10 μL of each nucleic acid and mix evenly (concentrations: 1.16×10 4 copies / μL, 1.62×10 4 copies / μL, 1.02×10 4 copies / μL, 1.94×10 4 copies / μL, 1.52×10 4 copies / μL) after serial dilution of 10 times, 100 times, 1000 times and 10000 times, then used as a template for five-fold fluorescent quantitative PCR detection, each gradient was repeated 3 times, and the minimum detection limit and linearity of the five-fold fluorescent quantitative PCR method were calculated. relation. Amplification results such as Image 6 As shown in Table 11, ASFV is the standard curve of the Texas Red channel (No. 1-Texas Red) regression equation is Y= -3.251X+40.547, and the correlation coefficient (R 2 ) was 0.990, the a...
Embodiment 3
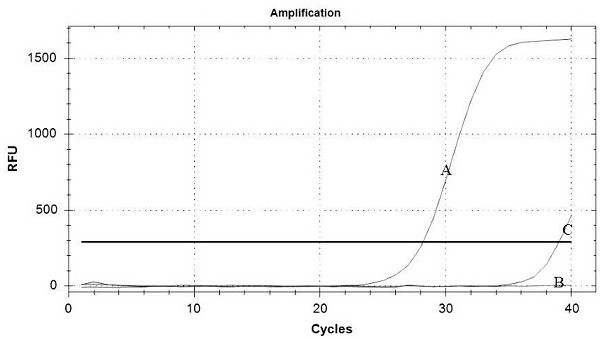
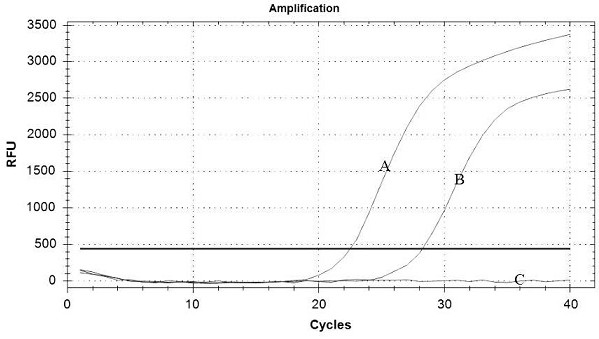
[0131] Embodiment 3: Five-fold fluorescent quantitative PCR detection method specific verification
[0132] FMDV (A type, O type, Asian type I), PRRSV (LV strain, JXA1 strain, VR2332 strain), CSFV (C strain), PRV standard strain, PRV vaccine strain (Bartha -K61 strain), TGEV-PEDV - PoRV was extracted according to step 3 of Example 1, and then the nucleic acid of the ASFV positive sample was used as a template for five-fold fluorescent quantitative PCR detection.
[0133] Test results such as Figure 7 As shown, except ASFV (No. 1), FMDV-A (No. 2), FMDV-O (No. 3), FMDV-Asia Ⅰ (No. 4), PRRSV-JXA1 (No. 5), CSFV (No. 6), PRV Except for wild virus (No. 7), other viruses (No. 8-10) were not detected, indicating that this method has no cross-reaction with other viruses, and can specifically detect ASFV, FMDV, HP-PRRSV, CSFV and PRV wild poison.
PUM
| Property | Measurement | Unit |
|---|---|---|
| correlation coefficient | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com